
Antarctic penguin virus A
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Avulavirinae; Orthoavulavirus; Avian orthoavulavirus 17
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
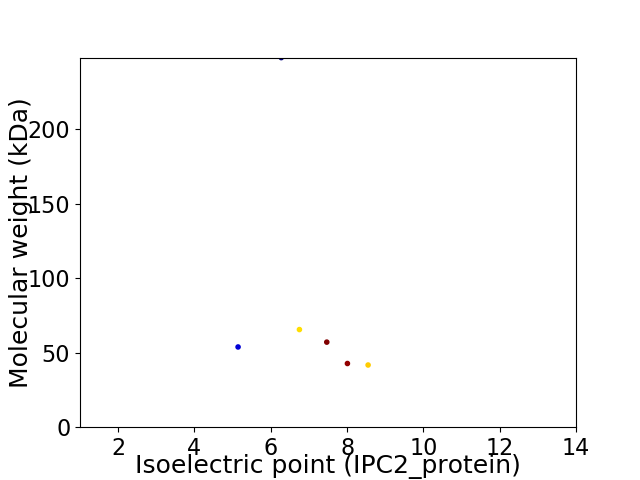
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y0KBV0|A0A1Y0KBV0_9MONO Phosphoprotein OS=Antarctic penguin virus A OX=2006072 PE=4 SV=1
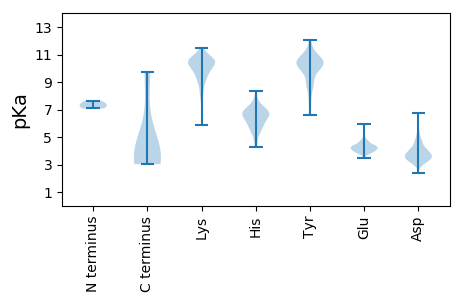
MM1 pKa = 7.33SSVFAEE7 pKa = 3.64YY8 pKa = 10.93DD9 pKa = 3.23KK10 pKa = 11.26FLEE13 pKa = 4.62NQTHH17 pKa = 6.01PPSRR21 pKa = 11.84ALPTEE26 pKa = 4.44GGGSLKK32 pKa = 10.74VEE34 pKa = 4.0VPVFVLNSDD43 pKa = 3.78DD44 pKa = 4.6PEE46 pKa = 4.24LRR48 pKa = 11.84WRR50 pKa = 11.84FVCFCLRR57 pKa = 11.84LAISEE62 pKa = 4.23SSNRR66 pKa = 11.84PLRR69 pKa = 11.84QGALISVLCAHH80 pKa = 6.56AQVMKK85 pKa = 10.27TYY87 pKa = 9.07ATLAAQSGEE96 pKa = 3.92AVITILEE103 pKa = 4.32IDD105 pKa = 4.38DD106 pKa = 5.65FDD108 pKa = 6.77DD109 pKa = 4.4KK110 pKa = 11.44MPVFNSRR117 pKa = 11.84SGITDD122 pKa = 2.93EE123 pKa = 4.27RR124 pKa = 11.84AARR127 pKa = 11.84LALIAADD134 pKa = 3.67IPRR137 pKa = 11.84ACSNEE142 pKa = 4.11TPFANAATEE151 pKa = 4.03NEE153 pKa = 4.21PPEE156 pKa = 4.66DD157 pKa = 4.57ASDD160 pKa = 3.45TLDD163 pKa = 4.1RR164 pKa = 11.84IFSVQVQVWITVAKK178 pKa = 10.69AMTAFEE184 pKa = 4.14TAEE187 pKa = 3.87EE188 pKa = 4.62SEE190 pKa = 4.11TRR192 pKa = 11.84RR193 pKa = 11.84LNKK196 pKa = 9.94YY197 pKa = 6.6MQQGRR202 pKa = 11.84VQKK205 pKa = 10.59KK206 pKa = 8.44CLLYY210 pKa = 10.01PIVRR214 pKa = 11.84TTVQMTIRR222 pKa = 11.84QSLVIRR228 pKa = 11.84GFLVNEE234 pKa = 4.39MKK236 pKa = 10.63RR237 pKa = 11.84AQNSPGGKK245 pKa = 7.92SAYY248 pKa = 9.76YY249 pKa = 10.5SFVGDD254 pKa = 3.49ISAYY258 pKa = 9.24IKK260 pKa = 10.75NAGVTAFLLTLKK272 pKa = 10.75FGIQTRR278 pKa = 11.84LPALALSSLAGDD290 pKa = 3.52IQKK293 pKa = 10.13VKK295 pKa = 10.86QLMVLYY301 pKa = 10.29RR302 pKa = 11.84EE303 pKa = 4.35KK304 pKa = 11.31GEE306 pKa = 4.38NGPFMTLLGDD316 pKa = 3.88PDD318 pKa = 3.59QMQFAPAEE326 pKa = 4.01YY327 pKa = 10.29SLLYY331 pKa = 10.23SYY333 pKa = 11.85AMGVASVLEE342 pKa = 4.46ASTSRR347 pKa = 11.84YY348 pKa = 8.5QFARR352 pKa = 11.84DD353 pKa = 3.51FMNPTFWRR361 pKa = 11.84IGVEE365 pKa = 4.3SAQSLSTSVDD375 pKa = 2.96EE376 pKa = 5.23QMATEE381 pKa = 4.25LQLGRR386 pKa = 11.84QSRR389 pKa = 11.84AALGEE394 pKa = 3.94MMAKK398 pKa = 9.69VAGSAGEE405 pKa = 4.06YY406 pKa = 9.77TMNAPAAAVMIGASGPNPPSSARR429 pKa = 11.84QSGSGPKK436 pKa = 8.23QTEE439 pKa = 3.85SDD441 pKa = 3.26IQLPPGFNTPEE452 pKa = 4.78DD453 pKa = 3.58YY454 pKa = 10.89LAYY457 pKa = 9.73IRR459 pKa = 11.84NEE461 pKa = 3.86EE462 pKa = 3.98AFKK465 pKa = 10.81SGRR468 pKa = 11.84ATSSTEE474 pKa = 3.91VPKK477 pKa = 10.8TPGLAGDD484 pKa = 4.77NDD486 pKa = 4.1PQVDD490 pKa = 3.58WEE492 pKa = 4.29LL493 pKa = 4.01
MM1 pKa = 7.33SSVFAEE7 pKa = 3.64YY8 pKa = 10.93DD9 pKa = 3.23KK10 pKa = 11.26FLEE13 pKa = 4.62NQTHH17 pKa = 6.01PPSRR21 pKa = 11.84ALPTEE26 pKa = 4.44GGGSLKK32 pKa = 10.74VEE34 pKa = 4.0VPVFVLNSDD43 pKa = 3.78DD44 pKa = 4.6PEE46 pKa = 4.24LRR48 pKa = 11.84WRR50 pKa = 11.84FVCFCLRR57 pKa = 11.84LAISEE62 pKa = 4.23SSNRR66 pKa = 11.84PLRR69 pKa = 11.84QGALISVLCAHH80 pKa = 6.56AQVMKK85 pKa = 10.27TYY87 pKa = 9.07ATLAAQSGEE96 pKa = 3.92AVITILEE103 pKa = 4.32IDD105 pKa = 4.38DD106 pKa = 5.65FDD108 pKa = 6.77DD109 pKa = 4.4KK110 pKa = 11.44MPVFNSRR117 pKa = 11.84SGITDD122 pKa = 2.93EE123 pKa = 4.27RR124 pKa = 11.84AARR127 pKa = 11.84LALIAADD134 pKa = 3.67IPRR137 pKa = 11.84ACSNEE142 pKa = 4.11TPFANAATEE151 pKa = 4.03NEE153 pKa = 4.21PPEE156 pKa = 4.66DD157 pKa = 4.57ASDD160 pKa = 3.45TLDD163 pKa = 4.1RR164 pKa = 11.84IFSVQVQVWITVAKK178 pKa = 10.69AMTAFEE184 pKa = 4.14TAEE187 pKa = 3.87EE188 pKa = 4.62SEE190 pKa = 4.11TRR192 pKa = 11.84RR193 pKa = 11.84LNKK196 pKa = 9.94YY197 pKa = 6.6MQQGRR202 pKa = 11.84VQKK205 pKa = 10.59KK206 pKa = 8.44CLLYY210 pKa = 10.01PIVRR214 pKa = 11.84TTVQMTIRR222 pKa = 11.84QSLVIRR228 pKa = 11.84GFLVNEE234 pKa = 4.39MKK236 pKa = 10.63RR237 pKa = 11.84AQNSPGGKK245 pKa = 7.92SAYY248 pKa = 9.76YY249 pKa = 10.5SFVGDD254 pKa = 3.49ISAYY258 pKa = 9.24IKK260 pKa = 10.75NAGVTAFLLTLKK272 pKa = 10.75FGIQTRR278 pKa = 11.84LPALALSSLAGDD290 pKa = 3.52IQKK293 pKa = 10.13VKK295 pKa = 10.86QLMVLYY301 pKa = 10.29RR302 pKa = 11.84EE303 pKa = 4.35KK304 pKa = 11.31GEE306 pKa = 4.38NGPFMTLLGDD316 pKa = 3.88PDD318 pKa = 3.59QMQFAPAEE326 pKa = 4.01YY327 pKa = 10.29SLLYY331 pKa = 10.23SYY333 pKa = 11.85AMGVASVLEE342 pKa = 4.46ASTSRR347 pKa = 11.84YY348 pKa = 8.5QFARR352 pKa = 11.84DD353 pKa = 3.51FMNPTFWRR361 pKa = 11.84IGVEE365 pKa = 4.3SAQSLSTSVDD375 pKa = 2.96EE376 pKa = 5.23QMATEE381 pKa = 4.25LQLGRR386 pKa = 11.84QSRR389 pKa = 11.84AALGEE394 pKa = 3.94MMAKK398 pKa = 9.69VAGSAGEE405 pKa = 4.06YY406 pKa = 9.77TMNAPAAAVMIGASGPNPPSSARR429 pKa = 11.84QSGSGPKK436 pKa = 8.23QTEE439 pKa = 3.85SDD441 pKa = 3.26IQLPPGFNTPEE452 pKa = 4.78DD453 pKa = 3.58YY454 pKa = 10.89LAYY457 pKa = 9.73IRR459 pKa = 11.84NEE461 pKa = 3.86EE462 pKa = 3.98AFKK465 pKa = 10.81SGRR468 pKa = 11.84ATSSTEE474 pKa = 3.91VPKK477 pKa = 10.8TPGLAGDD484 pKa = 4.77NDD486 pKa = 4.1PQVDD490 pKa = 3.58WEE492 pKa = 4.29LL493 pKa = 4.01
Molecular weight: 53.9 kDa
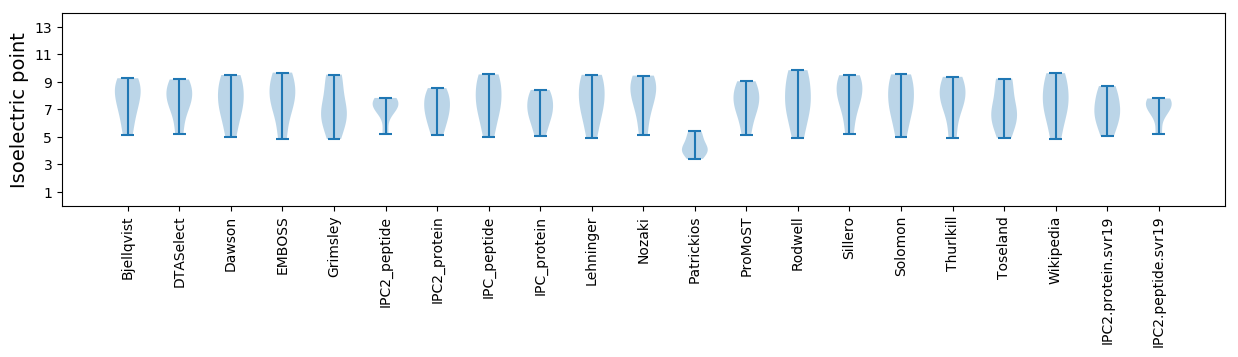
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y0KBW5|A0A1Y0KBW5_9MONO Hemagglutinin-neuraminidase OS=Antarctic penguin virus A OX=2006072 PE=3 SV=1
MM1 pKa = 7.51LRR3 pKa = 11.84AEE5 pKa = 4.79DD6 pKa = 3.68LASRR10 pKa = 11.84PIIGQQPAISMEE22 pKa = 3.84SRR24 pKa = 11.84TVKK27 pKa = 10.6LYY29 pKa = 11.02VDD31 pKa = 3.97PDD33 pKa = 3.76EE34 pKa = 4.99PASSLLAFPLIFTSTSDD51 pKa = 3.27GKK53 pKa = 9.75KK54 pKa = 8.33TLSPQYY60 pKa = 9.81RR61 pKa = 11.84IQIIDD66 pKa = 3.7GEE68 pKa = 4.42RR69 pKa = 11.84EE70 pKa = 3.9SDD72 pKa = 3.22SDD74 pKa = 4.76LIFISTYY81 pKa = 10.5GFISGMEE88 pKa = 4.01TAVDD92 pKa = 3.29KK93 pKa = 11.38SLAVDD98 pKa = 3.77MGQEE102 pKa = 3.99RR103 pKa = 11.84TILTSCMLPLGSIPKK118 pKa = 9.82VPDD121 pKa = 3.08FHH123 pKa = 8.33GLAKK127 pKa = 10.39SCLEE131 pKa = 4.39LKK133 pKa = 10.52VSCKK137 pKa = 10.03KK138 pKa = 10.51AATNSEE144 pKa = 4.1RR145 pKa = 11.84IIFNVTDD152 pKa = 4.48FPPILAQCAAIKK164 pKa = 10.55KK165 pKa = 10.07GVTSCNASVNLKK177 pKa = 10.49APEE180 pKa = 4.83KK181 pKa = 10.88IMGNYY186 pKa = 9.91DD187 pKa = 3.49LVYY190 pKa = 10.49KK191 pKa = 9.23VTFVSLTVIPASLVYY206 pKa = 10.18KK207 pKa = 10.52VSSPVLKK214 pKa = 10.57AGSSLTYY221 pKa = 9.72SLNMSVIIKK230 pKa = 10.25VDD232 pKa = 3.02ITDD235 pKa = 3.44KK236 pKa = 11.2HH237 pKa = 6.08PSAKK241 pKa = 10.39LLLKK245 pKa = 10.44KK246 pKa = 10.41DD247 pKa = 3.8DD248 pKa = 3.85QYY250 pKa = 12.08LANLWVHH257 pKa = 6.71WGLISAVKK265 pKa = 10.37KK266 pKa = 9.72GGKK269 pKa = 8.63RR270 pKa = 11.84HH271 pKa = 5.76TIEE274 pKa = 3.9EE275 pKa = 4.17VAEE278 pKa = 4.34KK279 pKa = 10.24IRR281 pKa = 11.84RR282 pKa = 11.84LDD284 pKa = 3.4IKK286 pKa = 10.78IEE288 pKa = 3.94LVDD291 pKa = 4.2LFGPSLIIQCKK302 pKa = 7.99GVKK305 pKa = 8.84TKK307 pKa = 10.8LLAGFFSKK315 pKa = 10.38QGTAVYY321 pKa = 8.82PISRR325 pKa = 11.84AAPQIGKK332 pKa = 9.15LLWSQTGTIMEE343 pKa = 4.57ASVVIQGGNQTQLASTSDD361 pKa = 3.79YY362 pKa = 10.95VIEE365 pKa = 4.29STKK368 pKa = 9.71VTLGKK373 pKa = 10.81GNSKK377 pKa = 10.61YY378 pKa = 11.05NPFRR382 pKa = 11.84KK383 pKa = 9.75
MM1 pKa = 7.51LRR3 pKa = 11.84AEE5 pKa = 4.79DD6 pKa = 3.68LASRR10 pKa = 11.84PIIGQQPAISMEE22 pKa = 3.84SRR24 pKa = 11.84TVKK27 pKa = 10.6LYY29 pKa = 11.02VDD31 pKa = 3.97PDD33 pKa = 3.76EE34 pKa = 4.99PASSLLAFPLIFTSTSDD51 pKa = 3.27GKK53 pKa = 9.75KK54 pKa = 8.33TLSPQYY60 pKa = 9.81RR61 pKa = 11.84IQIIDD66 pKa = 3.7GEE68 pKa = 4.42RR69 pKa = 11.84EE70 pKa = 3.9SDD72 pKa = 3.22SDD74 pKa = 4.76LIFISTYY81 pKa = 10.5GFISGMEE88 pKa = 4.01TAVDD92 pKa = 3.29KK93 pKa = 11.38SLAVDD98 pKa = 3.77MGQEE102 pKa = 3.99RR103 pKa = 11.84TILTSCMLPLGSIPKK118 pKa = 9.82VPDD121 pKa = 3.08FHH123 pKa = 8.33GLAKK127 pKa = 10.39SCLEE131 pKa = 4.39LKK133 pKa = 10.52VSCKK137 pKa = 10.03KK138 pKa = 10.51AATNSEE144 pKa = 4.1RR145 pKa = 11.84IIFNVTDD152 pKa = 4.48FPPILAQCAAIKK164 pKa = 10.55KK165 pKa = 10.07GVTSCNASVNLKK177 pKa = 10.49APEE180 pKa = 4.83KK181 pKa = 10.88IMGNYY186 pKa = 9.91DD187 pKa = 3.49LVYY190 pKa = 10.49KK191 pKa = 9.23VTFVSLTVIPASLVYY206 pKa = 10.18KK207 pKa = 10.52VSSPVLKK214 pKa = 10.57AGSSLTYY221 pKa = 9.72SLNMSVIIKK230 pKa = 10.25VDD232 pKa = 3.02ITDD235 pKa = 3.44KK236 pKa = 11.2HH237 pKa = 6.08PSAKK241 pKa = 10.39LLLKK245 pKa = 10.44KK246 pKa = 10.41DD247 pKa = 3.8DD248 pKa = 3.85QYY250 pKa = 12.08LANLWVHH257 pKa = 6.71WGLISAVKK265 pKa = 10.37KK266 pKa = 9.72GGKK269 pKa = 8.63RR270 pKa = 11.84HH271 pKa = 5.76TIEE274 pKa = 3.9EE275 pKa = 4.17VAEE278 pKa = 4.34KK279 pKa = 10.24IRR281 pKa = 11.84RR282 pKa = 11.84LDD284 pKa = 3.4IKK286 pKa = 10.78IEE288 pKa = 3.94LVDD291 pKa = 4.2LFGPSLIIQCKK302 pKa = 7.99GVKK305 pKa = 8.84TKK307 pKa = 10.8LLAGFFSKK315 pKa = 10.38QGTAVYY321 pKa = 8.82PISRR325 pKa = 11.84AAPQIGKK332 pKa = 9.15LLWSQTGTIMEE343 pKa = 4.57ASVVIQGGNQTQLASTSDD361 pKa = 3.79YY362 pKa = 10.95VIEE365 pKa = 4.29STKK368 pKa = 9.71VTLGKK373 pKa = 10.81GNSKK377 pKa = 10.61YY378 pKa = 11.05NPFRR382 pKa = 11.84KK383 pKa = 9.75
Molecular weight: 41.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4620 |
383 |
2202 |
770.0 |
84.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.835 ± 1.107 | 1.991 ± 0.372 |
5.173 ± 0.49 | 4.156 ± 0.417 |
3.247 ± 0.383 | 5.952 ± 0.513 |
1.797 ± 0.37 | 7.662 ± 0.72 |
4.372 ± 0.538 | 10.584 ± 0.879 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.532 ± 0.373 | 4.567 ± 0.345 |
4.87 ± 0.54 | 5.065 ± 0.514 |
5.043 ± 0.63 | 8.918 ± 0.525 |
6.212 ± 0.511 | 5.931 ± 0.438 |
0.844 ± 0.179 | 3.247 ± 0.475 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
