
Dendrothele bispora (strain CBS 962.96)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetidae; Agaricales; Agaricales incertae sedis; Dendrothele; Dendrothele bispora
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
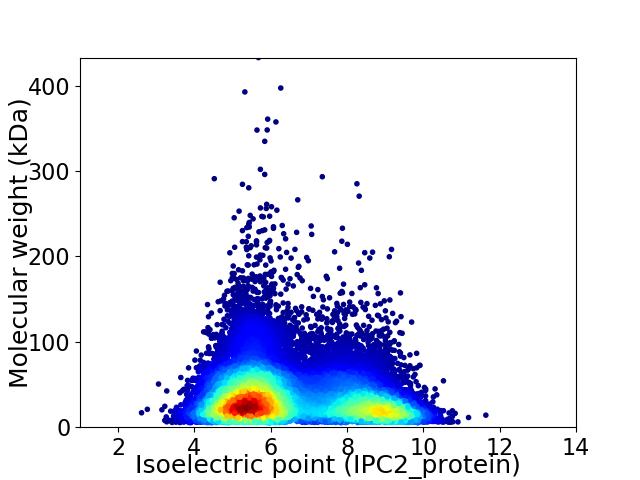
Virtual 2D-PAGE plot for 33018 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4S8LN97|A0A4S8LN97_DENBC HAD-like protein OS=Dendrothele bispora (strain CBS 962.96) OX=1314807 GN=K435DRAFT_759899 PE=4 SV=1
MM1 pKa = 7.66SSEE4 pKa = 4.16SSNLSSLTRR13 pKa = 11.84SKK15 pKa = 10.6LHH17 pKa = 6.54HH18 pKa = 6.44SVGSKK23 pKa = 10.22DD24 pKa = 3.13KK25 pKa = 10.95CSLHH29 pKa = 6.45RR30 pKa = 11.84WVLLKK35 pKa = 11.01NSFVRR40 pKa = 11.84ALPLTTNNTSPSNEE54 pKa = 3.79AMDD57 pKa = 4.59ANYY60 pKa = 10.63SEE62 pKa = 4.64SPRR65 pKa = 11.84GAEE68 pKa = 3.93EE69 pKa = 4.0TASDD73 pKa = 4.19DD74 pKa = 3.63ADD76 pKa = 3.39SFMFPDD82 pKa = 4.05VSNMAEE88 pKa = 3.94SSGVDD93 pKa = 3.32GGASEE98 pKa = 4.91AQWLDD103 pKa = 3.35SLLEE107 pKa = 4.05TLGDD111 pKa = 4.5DD112 pKa = 5.05DD113 pKa = 7.09DD114 pKa = 7.47DD115 pKa = 5.48DD116 pKa = 5.51FSVDD120 pKa = 3.48NVSVLQPDD128 pKa = 4.25DD129 pKa = 5.95DD130 pKa = 5.39DD131 pKa = 5.48DD132 pKa = 6.27HH133 pKa = 6.44MLSPIVSPMSSSDD146 pKa = 3.71DD147 pKa = 3.61LSSVDD152 pKa = 4.97FSSQADD158 pKa = 3.89FSSPIAVSYY167 pKa = 8.15PYY169 pKa = 9.6PVPYY173 pKa = 10.25PPFHH177 pKa = 7.19PPLISYY183 pKa = 8.34HH184 pKa = 5.7QLDD187 pKa = 3.93SSFDD191 pKa = 3.85PLDD194 pKa = 3.31ITGDD198 pKa = 3.7PLPYY202 pKa = 10.07YY203 pKa = 10.45DD204 pKa = 5.86LEE206 pKa = 4.56DD207 pKa = 4.43VEE209 pKa = 5.43DD210 pKa = 4.86LPVPDD215 pKa = 5.8AIEE218 pKa = 4.42DD219 pKa = 3.94TSDD222 pKa = 4.15DD223 pKa = 3.93EE224 pKa = 6.02SDD226 pKa = 3.56TLLTPSVGHH235 pKa = 6.29SSSSLLSLDD244 pKa = 3.91SSPAPLSADD253 pKa = 3.44RR254 pKa = 11.84SRR256 pKa = 11.84SHH258 pKa = 6.68HH259 pKa = 6.6SSPHH263 pKa = 6.61VYY265 pKa = 9.53IDD267 pKa = 3.72SDD269 pKa = 3.72DD270 pKa = 3.93SSFYY274 pKa = 10.46PYY276 pKa = 9.63EE277 pKa = 4.29TDD279 pKa = 3.67PDD281 pKa = 4.13PLPFSDD287 pKa = 5.13HH288 pKa = 6.43LRR290 pKa = 11.84SPYY293 pKa = 10.9NFYY296 pKa = 10.65QEE298 pKa = 4.38CC299 pKa = 3.57
MM1 pKa = 7.66SSEE4 pKa = 4.16SSNLSSLTRR13 pKa = 11.84SKK15 pKa = 10.6LHH17 pKa = 6.54HH18 pKa = 6.44SVGSKK23 pKa = 10.22DD24 pKa = 3.13KK25 pKa = 10.95CSLHH29 pKa = 6.45RR30 pKa = 11.84WVLLKK35 pKa = 11.01NSFVRR40 pKa = 11.84ALPLTTNNTSPSNEE54 pKa = 3.79AMDD57 pKa = 4.59ANYY60 pKa = 10.63SEE62 pKa = 4.64SPRR65 pKa = 11.84GAEE68 pKa = 3.93EE69 pKa = 4.0TASDD73 pKa = 4.19DD74 pKa = 3.63ADD76 pKa = 3.39SFMFPDD82 pKa = 4.05VSNMAEE88 pKa = 3.94SSGVDD93 pKa = 3.32GGASEE98 pKa = 4.91AQWLDD103 pKa = 3.35SLLEE107 pKa = 4.05TLGDD111 pKa = 4.5DD112 pKa = 5.05DD113 pKa = 7.09DD114 pKa = 7.47DD115 pKa = 5.48DD116 pKa = 5.51FSVDD120 pKa = 3.48NVSVLQPDD128 pKa = 4.25DD129 pKa = 5.95DD130 pKa = 5.39DD131 pKa = 5.48DD132 pKa = 6.27HH133 pKa = 6.44MLSPIVSPMSSSDD146 pKa = 3.71DD147 pKa = 3.61LSSVDD152 pKa = 4.97FSSQADD158 pKa = 3.89FSSPIAVSYY167 pKa = 8.15PYY169 pKa = 9.6PVPYY173 pKa = 10.25PPFHH177 pKa = 7.19PPLISYY183 pKa = 8.34HH184 pKa = 5.7QLDD187 pKa = 3.93SSFDD191 pKa = 3.85PLDD194 pKa = 3.31ITGDD198 pKa = 3.7PLPYY202 pKa = 10.07YY203 pKa = 10.45DD204 pKa = 5.86LEE206 pKa = 4.56DD207 pKa = 4.43VEE209 pKa = 5.43DD210 pKa = 4.86LPVPDD215 pKa = 5.8AIEE218 pKa = 4.42DD219 pKa = 3.94TSDD222 pKa = 4.15DD223 pKa = 3.93EE224 pKa = 6.02SDD226 pKa = 3.56TLLTPSVGHH235 pKa = 6.29SSSSLLSLDD244 pKa = 3.91SSPAPLSADD253 pKa = 3.44RR254 pKa = 11.84SRR256 pKa = 11.84SHH258 pKa = 6.68HH259 pKa = 6.6SSPHH263 pKa = 6.61VYY265 pKa = 9.53IDD267 pKa = 3.72SDD269 pKa = 3.72DD270 pKa = 3.93SSFYY274 pKa = 10.46PYY276 pKa = 9.63EE277 pKa = 4.29TDD279 pKa = 3.67PDD281 pKa = 4.13PLPFSDD287 pKa = 5.13HH288 pKa = 6.43LRR290 pKa = 11.84SPYY293 pKa = 10.9NFYY296 pKa = 10.65QEE298 pKa = 4.38CC299 pKa = 3.57
Molecular weight: 32.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4S8L737|A0A4S8L737_DENBC Uncharacterized protein (Fragment) OS=Dendrothele bispora (strain CBS 962.96) OX=1314807 GN=K435DRAFT_593729 PE=4 SV=1
MM1 pKa = 8.2RR2 pKa = 11.84IRR4 pKa = 11.84NQSMTSWPRR13 pKa = 11.84WSPRR17 pKa = 11.84RR18 pKa = 11.84PQPPSWSPRR27 pKa = 11.84RR28 pKa = 11.84TPPSPFSKK36 pKa = 10.03PQPQSLTSSSLLPLTSPSTPSSQSPRR62 pKa = 11.84TTPPSQTPSAWPTWPARR79 pKa = 11.84SPSAWQTPLKK89 pKa = 10.01PLSTRR94 pKa = 11.84SLTHH98 pKa = 5.88PTSSS102 pKa = 3.17
MM1 pKa = 8.2RR2 pKa = 11.84IRR4 pKa = 11.84NQSMTSWPRR13 pKa = 11.84WSPRR17 pKa = 11.84RR18 pKa = 11.84PQPPSWSPRR27 pKa = 11.84RR28 pKa = 11.84TPPSPFSKK36 pKa = 10.03PQPQSLTSSSLLPLTSPSTPSSQSPRR62 pKa = 11.84TTPPSQTPSAWPTWPARR79 pKa = 11.84SPSAWQTPLKK89 pKa = 10.01PLSTRR94 pKa = 11.84SLTHH98 pKa = 5.88PTSSS102 pKa = 3.17
Molecular weight: 11.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
11777400 |
49 |
5921 |
356.7 |
39.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.135 ± 0.013 | 1.356 ± 0.006 |
5.656 ± 0.016 | 6.036 ± 0.017 |
3.993 ± 0.009 | 6.401 ± 0.016 |
2.54 ± 0.008 | 5.147 ± 0.01 |
4.87 ± 0.013 | 9.123 ± 0.015 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.107 ± 0.005 | 4.011 ± 0.009 |
6.06 ± 0.016 | 3.841 ± 0.009 |
5.876 ± 0.011 | 9.134 ± 0.022 |
6.188 ± 0.011 | 6.252 ± 0.026 |
1.511 ± 0.005 | 2.763 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
