
human papillomavirus 118
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Betapapillomavirus; Betapapillomavirus 1
Average proteome isoelectric point is 5.89
Get precalculated fractions of proteins
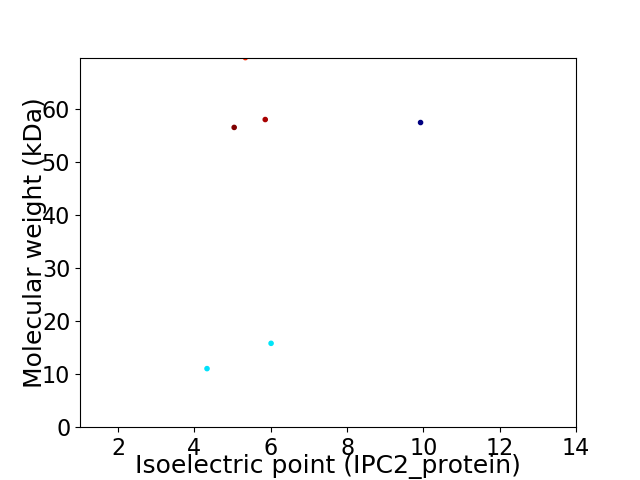
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
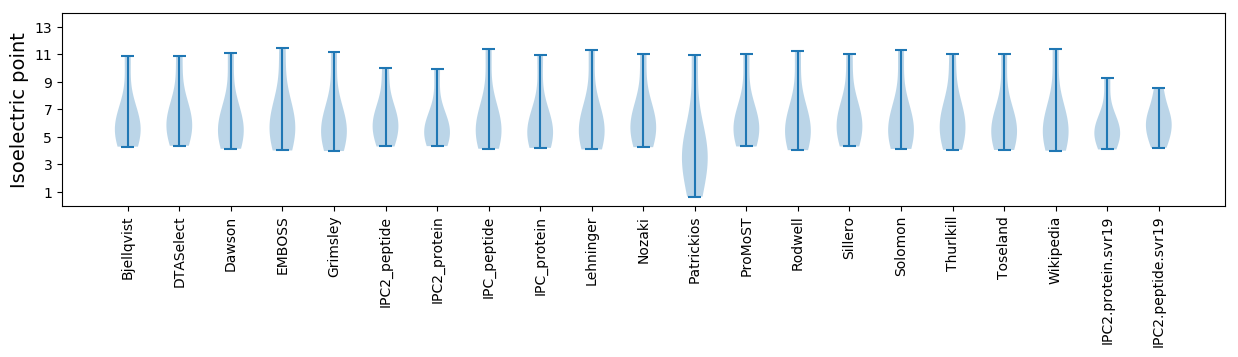
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D6MM12|D6MM12_9PAPI Replication protein E1 OS=human papillomavirus 118 OX=927771 GN=E1 PE=3 SV=1
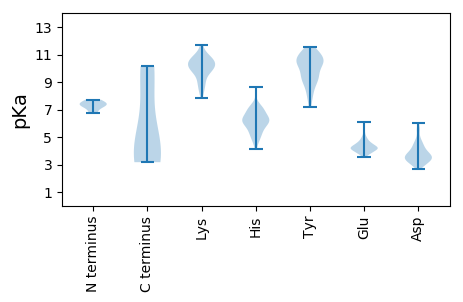
MM1 pKa = 7.18VGNEE5 pKa = 3.81VTLQDD10 pKa = 3.38IVLEE14 pKa = 4.57LEE16 pKa = 4.11PQAIDD21 pKa = 3.44LFCYY25 pKa = 10.38EE26 pKa = 4.69EE27 pKa = 4.17EE28 pKa = 4.97LPTEE32 pKa = 4.14QDD34 pKa = 2.97TSSEE38 pKa = 4.19EE39 pKa = 3.82EE40 pKa = 4.01PDD42 pKa = 3.7PVKK45 pKa = 10.61IPYY48 pKa = 9.65KK49 pKa = 10.38VVAPCGCCGPRR60 pKa = 11.84LRR62 pKa = 11.84LCLLATTFGIRR73 pKa = 11.84TLEE76 pKa = 3.95SLLLDD81 pKa = 5.34EE82 pKa = 6.1INLLCPPCRR91 pKa = 11.84HH92 pKa = 6.15SLQHH96 pKa = 6.44GNQQ99 pKa = 3.17
MM1 pKa = 7.18VGNEE5 pKa = 3.81VTLQDD10 pKa = 3.38IVLEE14 pKa = 4.57LEE16 pKa = 4.11PQAIDD21 pKa = 3.44LFCYY25 pKa = 10.38EE26 pKa = 4.69EE27 pKa = 4.17EE28 pKa = 4.97LPTEE32 pKa = 4.14QDD34 pKa = 2.97TSSEE38 pKa = 4.19EE39 pKa = 3.82EE40 pKa = 4.01PDD42 pKa = 3.7PVKK45 pKa = 10.61IPYY48 pKa = 9.65KK49 pKa = 10.38VVAPCGCCGPRR60 pKa = 11.84LRR62 pKa = 11.84LCLLATTFGIRR73 pKa = 11.84TLEE76 pKa = 3.95SLLLDD81 pKa = 5.34EE82 pKa = 6.1INLLCPPCRR91 pKa = 11.84HH92 pKa = 6.15SLQHH96 pKa = 6.44GNQQ99 pKa = 3.17
Molecular weight: 11.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D6MM14|D6MM14_9PAPI Minor capsid protein L2 OS=human papillomavirus 118 OX=927771 GN=L2 PE=3 SV=1
MM1 pKa = 7.41EE2 pKa = 5.52ALNEE6 pKa = 4.14RR7 pKa = 11.84FSALQDD13 pKa = 3.24QLMNIYY19 pKa = 9.44EE20 pKa = 4.32AAKK23 pKa = 9.2STLDD27 pKa = 3.33AQIDD31 pKa = 3.61HH32 pKa = 6.39WQHH35 pKa = 5.04LRR37 pKa = 11.84KK38 pKa = 9.43EE39 pKa = 4.16AVLLFVARR47 pKa = 11.84QKK49 pKa = 11.28GILRR53 pKa = 11.84LGYY56 pKa = 10.26QPVPPQAVSEE66 pKa = 4.52SKK68 pKa = 10.83AKK70 pKa = 9.84NAIMMVLQLEE80 pKa = 4.46SLKK83 pKa = 10.64KK84 pKa = 10.09SQFAEE89 pKa = 4.53EE90 pKa = 4.36PWTLVDD96 pKa = 4.68TSLEE100 pKa = 4.36TYY102 pKa = 10.62KK103 pKa = 10.73NAPEE107 pKa = 3.94NTFKK111 pKa = 10.92KK112 pKa = 10.62GPINVEE118 pKa = 3.55VMYY121 pKa = 11.01DD122 pKa = 3.61KK123 pKa = 11.49DD124 pKa = 3.73PDD126 pKa = 3.68NTNLYY131 pKa = 9.13TMWKK135 pKa = 8.59YY136 pKa = 10.71IYY138 pKa = 10.41YY139 pKa = 10.38LDD141 pKa = 5.73AEE143 pKa = 4.66EE144 pKa = 4.52EE145 pKa = 4.2WQKK148 pKa = 11.68AEE150 pKa = 4.29SGANHH155 pKa = 5.94TGIYY159 pKa = 9.4YY160 pKa = 7.56MHH162 pKa = 7.19GSFRR166 pKa = 11.84HH167 pKa = 5.42YY168 pKa = 10.68YY169 pKa = 10.53VLFADD174 pKa = 4.18DD175 pKa = 4.35APRR178 pKa = 11.84FSRR181 pKa = 11.84TGHH184 pKa = 5.74WEE186 pKa = 3.89VIINKK191 pKa = 8.54DD192 pKa = 3.45TVFAPVTSSTPPEE205 pKa = 4.24SPGQSQGRR213 pKa = 11.84QPPDD217 pKa = 2.99KK218 pKa = 10.12GTGITTTDD226 pKa = 3.08AATNRR231 pKa = 11.84SPRR234 pKa = 11.84ASPTTTAVTEE244 pKa = 4.29RR245 pKa = 11.84PQKK248 pKa = 10.17RR249 pKa = 11.84RR250 pKa = 11.84YY251 pKa = 8.45GRR253 pKa = 11.84KK254 pKa = 9.58DD255 pKa = 3.21SSPTATPKK263 pKa = 10.31KK264 pKa = 10.02RR265 pKa = 11.84STRR268 pKa = 11.84QTWQRR273 pKa = 11.84RR274 pKa = 11.84RR275 pKa = 11.84PRR277 pKa = 11.84SRR279 pKa = 11.84SLRR282 pKa = 11.84AEE284 pKa = 3.9KK285 pKa = 10.27RR286 pKa = 11.84RR287 pKa = 11.84QTRR290 pKa = 11.84SRR292 pKa = 11.84STSSKK297 pKa = 8.95ASKK300 pKa = 9.69NRR302 pKa = 11.84RR303 pKa = 11.84RR304 pKa = 11.84RR305 pKa = 11.84SSSSSSSSSCSRR317 pKa = 11.84GSSSRR322 pKa = 11.84RR323 pKa = 11.84RR324 pKa = 11.84RR325 pKa = 11.84RR326 pKa = 11.84TRR328 pKa = 11.84SRR330 pKa = 11.84SASRR334 pKa = 11.84TKK336 pKa = 10.78SPASRR341 pKa = 11.84TRR343 pKa = 11.84SRR345 pKa = 11.84TRR347 pKa = 11.84SPTSKK352 pKa = 10.03AQSPGRR358 pKa = 11.84RR359 pKa = 11.84AGGRR363 pKa = 11.84SSRR366 pKa = 11.84RR367 pKa = 11.84STSSSTSPSKK377 pKa = 9.88RR378 pKa = 11.84RR379 pKa = 11.84KK380 pKa = 8.37RR381 pKa = 11.84ARR383 pKa = 11.84DD384 pKa = 3.19RR385 pKa = 11.84GPSYY389 pKa = 11.29NRR391 pKa = 11.84GISPGDD397 pKa = 3.28VGKK400 pKa = 10.58SVQTVSGRR408 pKa = 11.84HH409 pKa = 5.26KK410 pKa = 10.87GRR412 pKa = 11.84LGRR415 pKa = 11.84LLAEE419 pKa = 4.16AHH421 pKa = 6.97DD422 pKa = 4.36PPVILVKK429 pKa = 10.75GDD431 pKa = 3.86PNTLKK436 pKa = 10.73CFRR439 pKa = 11.84NRR441 pKa = 11.84SKK443 pKa = 9.95TKK445 pKa = 10.16YY446 pKa = 9.29RR447 pKa = 11.84DD448 pKa = 3.39LIKK451 pKa = 10.6AVSTTWSWVSTDD463 pKa = 2.65GCDD466 pKa = 3.32RR467 pKa = 11.84IGRR470 pKa = 11.84ARR472 pKa = 11.84MLFSFASYY480 pKa = 8.17EE481 pKa = 4.02QRR483 pKa = 11.84NTFDD487 pKa = 3.96NKK489 pKa = 8.19VTYY492 pKa = 9.03PKK494 pKa = 10.54NVEE497 pKa = 3.88RR498 pKa = 11.84TFGNFDD504 pKa = 3.51SLL506 pKa = 4.21
MM1 pKa = 7.41EE2 pKa = 5.52ALNEE6 pKa = 4.14RR7 pKa = 11.84FSALQDD13 pKa = 3.24QLMNIYY19 pKa = 9.44EE20 pKa = 4.32AAKK23 pKa = 9.2STLDD27 pKa = 3.33AQIDD31 pKa = 3.61HH32 pKa = 6.39WQHH35 pKa = 5.04LRR37 pKa = 11.84KK38 pKa = 9.43EE39 pKa = 4.16AVLLFVARR47 pKa = 11.84QKK49 pKa = 11.28GILRR53 pKa = 11.84LGYY56 pKa = 10.26QPVPPQAVSEE66 pKa = 4.52SKK68 pKa = 10.83AKK70 pKa = 9.84NAIMMVLQLEE80 pKa = 4.46SLKK83 pKa = 10.64KK84 pKa = 10.09SQFAEE89 pKa = 4.53EE90 pKa = 4.36PWTLVDD96 pKa = 4.68TSLEE100 pKa = 4.36TYY102 pKa = 10.62KK103 pKa = 10.73NAPEE107 pKa = 3.94NTFKK111 pKa = 10.92KK112 pKa = 10.62GPINVEE118 pKa = 3.55VMYY121 pKa = 11.01DD122 pKa = 3.61KK123 pKa = 11.49DD124 pKa = 3.73PDD126 pKa = 3.68NTNLYY131 pKa = 9.13TMWKK135 pKa = 8.59YY136 pKa = 10.71IYY138 pKa = 10.41YY139 pKa = 10.38LDD141 pKa = 5.73AEE143 pKa = 4.66EE144 pKa = 4.52EE145 pKa = 4.2WQKK148 pKa = 11.68AEE150 pKa = 4.29SGANHH155 pKa = 5.94TGIYY159 pKa = 9.4YY160 pKa = 7.56MHH162 pKa = 7.19GSFRR166 pKa = 11.84HH167 pKa = 5.42YY168 pKa = 10.68YY169 pKa = 10.53VLFADD174 pKa = 4.18DD175 pKa = 4.35APRR178 pKa = 11.84FSRR181 pKa = 11.84TGHH184 pKa = 5.74WEE186 pKa = 3.89VIINKK191 pKa = 8.54DD192 pKa = 3.45TVFAPVTSSTPPEE205 pKa = 4.24SPGQSQGRR213 pKa = 11.84QPPDD217 pKa = 2.99KK218 pKa = 10.12GTGITTTDD226 pKa = 3.08AATNRR231 pKa = 11.84SPRR234 pKa = 11.84ASPTTTAVTEE244 pKa = 4.29RR245 pKa = 11.84PQKK248 pKa = 10.17RR249 pKa = 11.84RR250 pKa = 11.84YY251 pKa = 8.45GRR253 pKa = 11.84KK254 pKa = 9.58DD255 pKa = 3.21SSPTATPKK263 pKa = 10.31KK264 pKa = 10.02RR265 pKa = 11.84STRR268 pKa = 11.84QTWQRR273 pKa = 11.84RR274 pKa = 11.84RR275 pKa = 11.84PRR277 pKa = 11.84SRR279 pKa = 11.84SLRR282 pKa = 11.84AEE284 pKa = 3.9KK285 pKa = 10.27RR286 pKa = 11.84RR287 pKa = 11.84QTRR290 pKa = 11.84SRR292 pKa = 11.84STSSKK297 pKa = 8.95ASKK300 pKa = 9.69NRR302 pKa = 11.84RR303 pKa = 11.84RR304 pKa = 11.84RR305 pKa = 11.84SSSSSSSSSCSRR317 pKa = 11.84GSSSRR322 pKa = 11.84RR323 pKa = 11.84RR324 pKa = 11.84RR325 pKa = 11.84RR326 pKa = 11.84TRR328 pKa = 11.84SRR330 pKa = 11.84SASRR334 pKa = 11.84TKK336 pKa = 10.78SPASRR341 pKa = 11.84TRR343 pKa = 11.84SRR345 pKa = 11.84TRR347 pKa = 11.84SPTSKK352 pKa = 10.03AQSPGRR358 pKa = 11.84RR359 pKa = 11.84AGGRR363 pKa = 11.84SSRR366 pKa = 11.84RR367 pKa = 11.84STSSSTSPSKK377 pKa = 9.88RR378 pKa = 11.84RR379 pKa = 11.84KK380 pKa = 8.37RR381 pKa = 11.84ARR383 pKa = 11.84DD384 pKa = 3.19RR385 pKa = 11.84GPSYY389 pKa = 11.29NRR391 pKa = 11.84GISPGDD397 pKa = 3.28VGKK400 pKa = 10.58SVQTVSGRR408 pKa = 11.84HH409 pKa = 5.26KK410 pKa = 10.87GRR412 pKa = 11.84LGRR415 pKa = 11.84LLAEE419 pKa = 4.16AHH421 pKa = 6.97DD422 pKa = 4.36PPVILVKK429 pKa = 10.75GDD431 pKa = 3.86PNTLKK436 pKa = 10.73CFRR439 pKa = 11.84NRR441 pKa = 11.84SKK443 pKa = 9.95TKK445 pKa = 10.16YY446 pKa = 9.29RR447 pKa = 11.84DD448 pKa = 3.39LIKK451 pKa = 10.6AVSTTWSWVSTDD463 pKa = 2.65GCDD466 pKa = 3.32RR467 pKa = 11.84IGRR470 pKa = 11.84ARR472 pKa = 11.84MLFSFASYY480 pKa = 8.17EE481 pKa = 4.02QRR483 pKa = 11.84NTFDD487 pKa = 3.96NKK489 pKa = 8.19VTYY492 pKa = 9.03PKK494 pKa = 10.54NVEE497 pKa = 3.88RR498 pKa = 11.84TFGNFDD504 pKa = 3.51SLL506 pKa = 4.21
Molecular weight: 57.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2375 |
99 |
606 |
395.8 |
44.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.937 ± 0.555 | 2.105 ± 0.875 |
5.768 ± 0.49 | 6.274 ± 0.643 |
4.295 ± 0.561 | 5.768 ± 0.733 |
2.189 ± 0.236 | 5.011 ± 0.6 |
4.884 ± 0.975 | 7.916 ± 0.891 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.432 ± 0.303 | 4.421 ± 0.672 |
5.979 ± 0.873 | 4.968 ± 0.382 |
7.284 ± 1.292 | 8.295 ± 1.319 |
6.863 ± 1.147 | 5.895 ± 0.853 |
1.263 ± 0.382 | 3.453 ± 0.315 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
