
Hubei sobemo-like virus 19
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.26
Get precalculated fractions of proteins
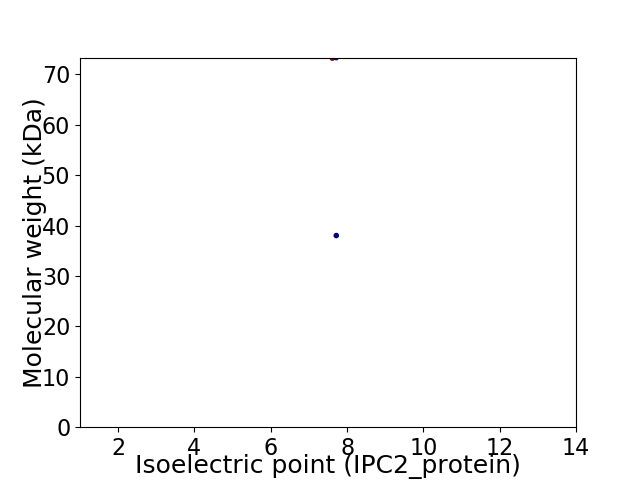
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
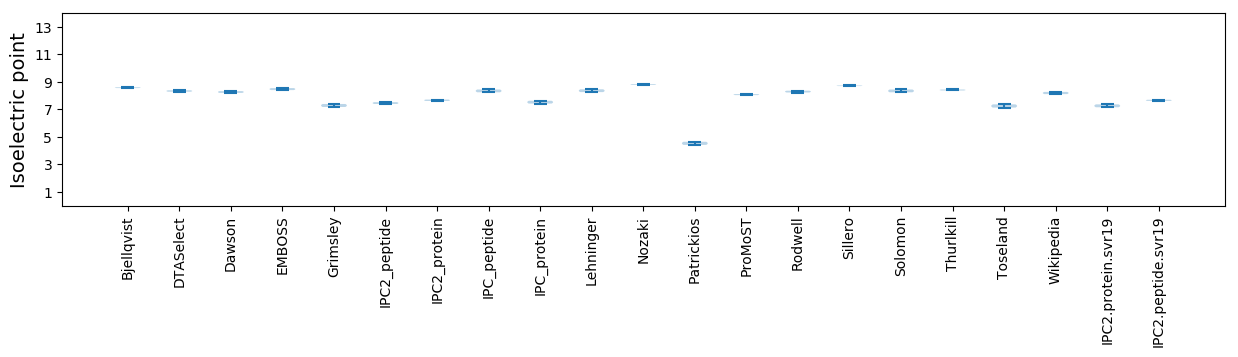
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEK7|A0A1L3KEK7_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 19 OX=1923204 PE=4 SV=1
MM1 pKa = 7.65EE2 pKa = 5.43PQPQEE7 pKa = 3.82NNFEE11 pKa = 4.15QEE13 pKa = 4.16EE14 pKa = 4.52LLVQQPILDD23 pKa = 4.15DD24 pKa = 4.07KK25 pKa = 11.12EE26 pKa = 4.74LNLFAKK32 pKa = 9.13MKK34 pKa = 9.17RR35 pKa = 11.84ICVITWRR42 pKa = 11.84SWLAIVHH49 pKa = 6.18LWLYY53 pKa = 10.7ISITKK58 pKa = 10.34YY59 pKa = 9.7LDD61 pKa = 3.02RR62 pKa = 11.84RR63 pKa = 11.84AEE65 pKa = 3.94EE66 pKa = 4.09LKK68 pKa = 10.01MEE70 pKa = 4.75EE71 pKa = 4.45KK72 pKa = 10.0MKK74 pKa = 10.54VWFPVFRR81 pKa = 11.84KK82 pKa = 9.13QLCFIYY88 pKa = 10.36NQLLVATQYY97 pKa = 11.19FDD99 pKa = 4.97AEE101 pKa = 4.69TFVLMLISALLSILTLWLFFKK122 pKa = 10.78KK123 pKa = 9.99SRR125 pKa = 11.84KK126 pKa = 7.34TLMSLRR132 pKa = 11.84GIKK135 pKa = 9.92FEE137 pKa = 4.1AMVEE141 pKa = 4.18GSHH144 pKa = 6.11FVKK147 pKa = 10.76GDD149 pKa = 3.11IPKK152 pKa = 9.97YY153 pKa = 9.92QVAILKK159 pKa = 10.48AGLLRR164 pKa = 11.84DD165 pKa = 3.45VHH167 pKa = 7.82VGFGIRR173 pKa = 11.84VNDD176 pKa = 3.77FLVVPSHH183 pKa = 7.1VINEE187 pKa = 4.16VGNDD191 pKa = 3.59LLLKK195 pKa = 10.63KK196 pKa = 10.25NGKK199 pKa = 7.95IQVSIGQRR207 pKa = 11.84IISDD211 pKa = 3.92YY212 pKa = 11.34VSDD215 pKa = 4.52LAYY218 pKa = 10.81LPISSTVWASLGVTRR233 pKa = 11.84ANLAKK238 pKa = 10.5EE239 pKa = 4.25SLALAAMATVTGNEE253 pKa = 4.13GQTTGLLRR261 pKa = 11.84KK262 pKa = 9.41CAIPFFYY269 pKa = 9.47TYY271 pKa = 9.51TASTVPGMSGAAYY284 pKa = 9.75VSNNMVHH291 pKa = 6.89AVHH294 pKa = 6.53CGVVGEE300 pKa = 4.58HH301 pKa = 6.35NSAISTVVVLKK312 pKa = 10.4EE313 pKa = 3.67LSKK316 pKa = 10.68IMKK319 pKa = 9.64GEE321 pKa = 3.8AVAATSISSNIDD333 pKa = 3.5MQWMPDD339 pKa = 3.21IDD341 pKa = 4.93KK342 pKa = 10.57VWHH345 pKa = 7.09DD346 pKa = 5.2DD347 pKa = 3.38IFDD350 pKa = 3.81KK351 pKa = 11.26YY352 pKa = 11.34VEE354 pKa = 4.24EE355 pKa = 4.63KK356 pKa = 10.78EE357 pKa = 4.14NRR359 pKa = 11.84LLNARR364 pKa = 11.84AKK366 pKa = 10.78HH367 pKa = 5.36GDD369 pKa = 3.28KK370 pKa = 10.99YY371 pKa = 11.35NPPEE375 pKa = 4.05QGWAAMMDD383 pKa = 3.68YY384 pKa = 11.17DD385 pKa = 5.76DD386 pKa = 6.18DD387 pKa = 3.95FAEE390 pKa = 4.84TYY392 pKa = 10.86GEE394 pKa = 4.18SFNVGKK400 pKa = 10.29NPRR403 pKa = 11.84SPMKK407 pKa = 10.71VRR409 pKa = 11.84ITPQGIIDD417 pKa = 4.23SDD419 pKa = 3.89SDD421 pKa = 3.14IDD423 pKa = 4.02VFVQRR428 pKa = 11.84TTDD431 pKa = 3.16AKK433 pKa = 8.37PTRR436 pKa = 11.84RR437 pKa = 11.84LEE439 pKa = 4.21LEE441 pKa = 3.9DD442 pKa = 5.88RR443 pKa = 11.84IANTEE448 pKa = 4.24DD449 pKa = 3.25GTLTNLHH456 pKa = 6.24LRR458 pKa = 11.84IKK460 pKa = 10.51RR461 pKa = 11.84LEE463 pKa = 4.0QKK465 pKa = 10.02VFQVDD470 pKa = 2.87KK471 pKa = 11.48CEE473 pKa = 3.8ICGAVTNQIAKK484 pKa = 9.97HH485 pKa = 5.4MKK487 pKa = 7.92TFHH490 pKa = 6.12GRR492 pKa = 11.84EE493 pKa = 4.1EE494 pKa = 4.27EE495 pKa = 4.1CWPCEE500 pKa = 4.01KK501 pKa = 10.67CSTVCKK507 pKa = 9.7TKK509 pKa = 10.76EE510 pKa = 3.64KK511 pKa = 10.9LEE513 pKa = 4.03RR514 pKa = 11.84HH515 pKa = 5.73LLSHH519 pKa = 6.67PEE521 pKa = 3.86RR522 pKa = 11.84PTCDD526 pKa = 2.59RR527 pKa = 11.84CGITCRR533 pKa = 11.84TQLTLLNHH541 pKa = 7.19RR542 pKa = 11.84NTCLPKK548 pKa = 10.07VAFNKK553 pKa = 8.87KK554 pKa = 6.24TEE556 pKa = 3.95KK557 pKa = 9.17VTLVGEE563 pKa = 4.33SAIPADD569 pKa = 4.05FKK571 pKa = 10.99TVVKK575 pKa = 10.44VDD577 pKa = 3.32KK578 pKa = 11.34NGFLGKK584 pKa = 10.22KK585 pKa = 10.1GSQKK589 pKa = 10.36KK590 pKa = 10.17DD591 pKa = 3.16SRR593 pKa = 11.84SSKK596 pKa = 7.57TTLNLSGEE604 pKa = 4.22KK605 pKa = 10.49DD606 pKa = 3.37PLTSQLEE613 pKa = 4.48FQSEE617 pKa = 4.37MLKK620 pKa = 10.25SQSAMLKK627 pKa = 10.13LLRR630 pKa = 11.84DD631 pKa = 3.96LAQNMNGQNLDD642 pKa = 2.9IMQNN646 pKa = 3.29
MM1 pKa = 7.65EE2 pKa = 5.43PQPQEE7 pKa = 3.82NNFEE11 pKa = 4.15QEE13 pKa = 4.16EE14 pKa = 4.52LLVQQPILDD23 pKa = 4.15DD24 pKa = 4.07KK25 pKa = 11.12EE26 pKa = 4.74LNLFAKK32 pKa = 9.13MKK34 pKa = 9.17RR35 pKa = 11.84ICVITWRR42 pKa = 11.84SWLAIVHH49 pKa = 6.18LWLYY53 pKa = 10.7ISITKK58 pKa = 10.34YY59 pKa = 9.7LDD61 pKa = 3.02RR62 pKa = 11.84RR63 pKa = 11.84AEE65 pKa = 3.94EE66 pKa = 4.09LKK68 pKa = 10.01MEE70 pKa = 4.75EE71 pKa = 4.45KK72 pKa = 10.0MKK74 pKa = 10.54VWFPVFRR81 pKa = 11.84KK82 pKa = 9.13QLCFIYY88 pKa = 10.36NQLLVATQYY97 pKa = 11.19FDD99 pKa = 4.97AEE101 pKa = 4.69TFVLMLISALLSILTLWLFFKK122 pKa = 10.78KK123 pKa = 9.99SRR125 pKa = 11.84KK126 pKa = 7.34TLMSLRR132 pKa = 11.84GIKK135 pKa = 9.92FEE137 pKa = 4.1AMVEE141 pKa = 4.18GSHH144 pKa = 6.11FVKK147 pKa = 10.76GDD149 pKa = 3.11IPKK152 pKa = 9.97YY153 pKa = 9.92QVAILKK159 pKa = 10.48AGLLRR164 pKa = 11.84DD165 pKa = 3.45VHH167 pKa = 7.82VGFGIRR173 pKa = 11.84VNDD176 pKa = 3.77FLVVPSHH183 pKa = 7.1VINEE187 pKa = 4.16VGNDD191 pKa = 3.59LLLKK195 pKa = 10.63KK196 pKa = 10.25NGKK199 pKa = 7.95IQVSIGQRR207 pKa = 11.84IISDD211 pKa = 3.92YY212 pKa = 11.34VSDD215 pKa = 4.52LAYY218 pKa = 10.81LPISSTVWASLGVTRR233 pKa = 11.84ANLAKK238 pKa = 10.5EE239 pKa = 4.25SLALAAMATVTGNEE253 pKa = 4.13GQTTGLLRR261 pKa = 11.84KK262 pKa = 9.41CAIPFFYY269 pKa = 9.47TYY271 pKa = 9.51TASTVPGMSGAAYY284 pKa = 9.75VSNNMVHH291 pKa = 6.89AVHH294 pKa = 6.53CGVVGEE300 pKa = 4.58HH301 pKa = 6.35NSAISTVVVLKK312 pKa = 10.4EE313 pKa = 3.67LSKK316 pKa = 10.68IMKK319 pKa = 9.64GEE321 pKa = 3.8AVAATSISSNIDD333 pKa = 3.5MQWMPDD339 pKa = 3.21IDD341 pKa = 4.93KK342 pKa = 10.57VWHH345 pKa = 7.09DD346 pKa = 5.2DD347 pKa = 3.38IFDD350 pKa = 3.81KK351 pKa = 11.26YY352 pKa = 11.34VEE354 pKa = 4.24EE355 pKa = 4.63KK356 pKa = 10.78EE357 pKa = 4.14NRR359 pKa = 11.84LLNARR364 pKa = 11.84AKK366 pKa = 10.78HH367 pKa = 5.36GDD369 pKa = 3.28KK370 pKa = 10.99YY371 pKa = 11.35NPPEE375 pKa = 4.05QGWAAMMDD383 pKa = 3.68YY384 pKa = 11.17DD385 pKa = 5.76DD386 pKa = 6.18DD387 pKa = 3.95FAEE390 pKa = 4.84TYY392 pKa = 10.86GEE394 pKa = 4.18SFNVGKK400 pKa = 10.29NPRR403 pKa = 11.84SPMKK407 pKa = 10.71VRR409 pKa = 11.84ITPQGIIDD417 pKa = 4.23SDD419 pKa = 3.89SDD421 pKa = 3.14IDD423 pKa = 4.02VFVQRR428 pKa = 11.84TTDD431 pKa = 3.16AKK433 pKa = 8.37PTRR436 pKa = 11.84RR437 pKa = 11.84LEE439 pKa = 4.21LEE441 pKa = 3.9DD442 pKa = 5.88RR443 pKa = 11.84IANTEE448 pKa = 4.24DD449 pKa = 3.25GTLTNLHH456 pKa = 6.24LRR458 pKa = 11.84IKK460 pKa = 10.51RR461 pKa = 11.84LEE463 pKa = 4.0QKK465 pKa = 10.02VFQVDD470 pKa = 2.87KK471 pKa = 11.48CEE473 pKa = 3.8ICGAVTNQIAKK484 pKa = 9.97HH485 pKa = 5.4MKK487 pKa = 7.92TFHH490 pKa = 6.12GRR492 pKa = 11.84EE493 pKa = 4.1EE494 pKa = 4.27EE495 pKa = 4.1CWPCEE500 pKa = 4.01KK501 pKa = 10.67CSTVCKK507 pKa = 9.7TKK509 pKa = 10.76EE510 pKa = 3.64KK511 pKa = 10.9LEE513 pKa = 4.03RR514 pKa = 11.84HH515 pKa = 5.73LLSHH519 pKa = 6.67PEE521 pKa = 3.86RR522 pKa = 11.84PTCDD526 pKa = 2.59RR527 pKa = 11.84CGITCRR533 pKa = 11.84TQLTLLNHH541 pKa = 7.19RR542 pKa = 11.84NTCLPKK548 pKa = 10.07VAFNKK553 pKa = 8.87KK554 pKa = 6.24TEE556 pKa = 3.95KK557 pKa = 9.17VTLVGEE563 pKa = 4.33SAIPADD569 pKa = 4.05FKK571 pKa = 10.99TVVKK575 pKa = 10.44VDD577 pKa = 3.32KK578 pKa = 11.34NGFLGKK584 pKa = 10.22KK585 pKa = 10.1GSQKK589 pKa = 10.36KK590 pKa = 10.17DD591 pKa = 3.16SRR593 pKa = 11.84SSKK596 pKa = 7.57TTLNLSGEE604 pKa = 4.22KK605 pKa = 10.49DD606 pKa = 3.37PLTSQLEE613 pKa = 4.48FQSEE617 pKa = 4.37MLKK620 pKa = 10.25SQSAMLKK627 pKa = 10.13LLRR630 pKa = 11.84DD631 pKa = 3.96LAQNMNGQNLDD642 pKa = 2.9IMQNN646 pKa = 3.29
Molecular weight: 73.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEK7|A0A1L3KEK7_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 19 OX=1923204 PE=4 SV=1
MM1 pKa = 7.74RR2 pKa = 11.84GSTTNRR8 pKa = 11.84QLLCYY13 pKa = 9.84KK14 pKa = 10.4NSDD17 pKa = 3.33VFDD20 pKa = 4.02PQKK23 pKa = 10.28IDD25 pKa = 3.53RR26 pKa = 11.84MWEE29 pKa = 3.63IVRR32 pKa = 11.84QKK34 pKa = 10.84LGGSGPDD41 pKa = 3.7PIKK44 pKa = 10.65IFVKK48 pKa = 10.42PEE50 pKa = 3.42PHH52 pKa = 5.96SLKK55 pKa = 10.6KK56 pKa = 10.42ISLKK60 pKa = 10.13RR61 pKa = 11.84YY62 pKa = 8.44RR63 pKa = 11.84LISSVSLADD72 pKa = 3.44QLIDD76 pKa = 3.25HH77 pKa = 6.47MLFGEE82 pKa = 4.49MNDD85 pKa = 3.64KK86 pKa = 10.93LIAGWPYY93 pKa = 10.86LPSKK97 pKa = 10.31VGWSFIGGGWKK108 pKa = 10.43AMPKK112 pKa = 9.77GGSWLAIDD120 pKa = 4.19KK121 pKa = 10.86SSWDD125 pKa = 3.13WTVRR129 pKa = 11.84PWLFDD134 pKa = 3.62LVLEE138 pKa = 4.7ARR140 pKa = 11.84LGLCRR145 pKa = 11.84TRR147 pKa = 11.84GEE149 pKa = 4.48VRR151 pKa = 11.84DD152 pKa = 3.86RR153 pKa = 11.84WSKK156 pKa = 9.67LAVMRR161 pKa = 11.84YY162 pKa = 7.1QQLFGNPIFITSGGFLLKK180 pKa = 10.14QKK182 pKa = 10.63RR183 pKa = 11.84PGVQKK188 pKa = 10.37SGCVNTISDD197 pKa = 3.81NSMMQSLLHH206 pKa = 6.35IRR208 pKa = 11.84VCLEE212 pKa = 3.61TNQPIGVLFSMGDD225 pKa = 3.44DD226 pKa = 3.86TLQQAPPLLDD236 pKa = 4.06EE237 pKa = 4.52YY238 pKa = 11.39LATLSKK244 pKa = 10.97YY245 pKa = 10.73CIVKK249 pKa = 9.83QAGDD253 pKa = 3.6EE254 pKa = 4.33VEE256 pKa = 4.12FAGFRR261 pKa = 11.84FDD263 pKa = 4.28GMLVEE268 pKa = 4.43PVYY271 pKa = 10.58HH272 pKa = 6.79GKK274 pKa = 9.02HH275 pKa = 6.08AYY277 pKa = 9.77NLLHH281 pKa = 7.3ADD283 pKa = 3.9PNVLTDD289 pKa = 4.03LSKK292 pKa = 10.84AYY294 pKa = 9.24PLLYY298 pKa = 9.84HH299 pKa = 6.97RR300 pKa = 11.84SSNRR304 pKa = 11.84EE305 pKa = 3.16WFRR308 pKa = 11.84NLFLDD313 pKa = 3.44WHH315 pKa = 6.57GEE317 pKa = 4.28VPSLDD322 pKa = 4.0YY323 pKa = 11.15LDD325 pKa = 5.34CIYY328 pKa = 11.17DD329 pKa = 3.59GFEE332 pKa = 3.67
MM1 pKa = 7.74RR2 pKa = 11.84GSTTNRR8 pKa = 11.84QLLCYY13 pKa = 9.84KK14 pKa = 10.4NSDD17 pKa = 3.33VFDD20 pKa = 4.02PQKK23 pKa = 10.28IDD25 pKa = 3.53RR26 pKa = 11.84MWEE29 pKa = 3.63IVRR32 pKa = 11.84QKK34 pKa = 10.84LGGSGPDD41 pKa = 3.7PIKK44 pKa = 10.65IFVKK48 pKa = 10.42PEE50 pKa = 3.42PHH52 pKa = 5.96SLKK55 pKa = 10.6KK56 pKa = 10.42ISLKK60 pKa = 10.13RR61 pKa = 11.84YY62 pKa = 8.44RR63 pKa = 11.84LISSVSLADD72 pKa = 3.44QLIDD76 pKa = 3.25HH77 pKa = 6.47MLFGEE82 pKa = 4.49MNDD85 pKa = 3.64KK86 pKa = 10.93LIAGWPYY93 pKa = 10.86LPSKK97 pKa = 10.31VGWSFIGGGWKK108 pKa = 10.43AMPKK112 pKa = 9.77GGSWLAIDD120 pKa = 4.19KK121 pKa = 10.86SSWDD125 pKa = 3.13WTVRR129 pKa = 11.84PWLFDD134 pKa = 3.62LVLEE138 pKa = 4.7ARR140 pKa = 11.84LGLCRR145 pKa = 11.84TRR147 pKa = 11.84GEE149 pKa = 4.48VRR151 pKa = 11.84DD152 pKa = 3.86RR153 pKa = 11.84WSKK156 pKa = 9.67LAVMRR161 pKa = 11.84YY162 pKa = 7.1QQLFGNPIFITSGGFLLKK180 pKa = 10.14QKK182 pKa = 10.63RR183 pKa = 11.84PGVQKK188 pKa = 10.37SGCVNTISDD197 pKa = 3.81NSMMQSLLHH206 pKa = 6.35IRR208 pKa = 11.84VCLEE212 pKa = 3.61TNQPIGVLFSMGDD225 pKa = 3.44DD226 pKa = 3.86TLQQAPPLLDD236 pKa = 4.06EE237 pKa = 4.52YY238 pKa = 11.39LATLSKK244 pKa = 10.97YY245 pKa = 10.73CIVKK249 pKa = 9.83QAGDD253 pKa = 3.6EE254 pKa = 4.33VEE256 pKa = 4.12FAGFRR261 pKa = 11.84FDD263 pKa = 4.28GMLVEE268 pKa = 4.43PVYY271 pKa = 10.58HH272 pKa = 6.79GKK274 pKa = 9.02HH275 pKa = 6.08AYY277 pKa = 9.77NLLHH281 pKa = 7.3ADD283 pKa = 3.9PNVLTDD289 pKa = 4.03LSKK292 pKa = 10.84AYY294 pKa = 9.24PLLYY298 pKa = 9.84HH299 pKa = 6.97RR300 pKa = 11.84SSNRR304 pKa = 11.84EE305 pKa = 3.16WFRR308 pKa = 11.84NLFLDD313 pKa = 3.44WHH315 pKa = 6.57GEE317 pKa = 4.28VPSLDD322 pKa = 4.0YY323 pKa = 11.15LDD325 pKa = 5.34CIYY328 pKa = 11.17DD329 pKa = 3.59GFEE332 pKa = 3.67
Molecular weight: 38.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
978 |
332 |
646 |
489.0 |
55.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.419 ± 0.898 | 2.045 ± 0.142 |
6.033 ± 0.534 | 5.624 ± 1.02 |
4.09 ± 0.256 | 6.135 ± 1.193 |
2.352 ± 0.035 | 5.521 ± 0.239 |
7.771 ± 0.863 | 10.941 ± 0.841 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.17 ± 0.094 | 4.294 ± 0.586 |
4.09 ± 0.795 | 4.09 ± 0.104 |
5.01 ± 0.425 | 6.646 ± 0.528 |
5.112 ± 1.254 | 6.851 ± 0.673 |
2.147 ± 0.696 | 2.658 ± 0.571 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
