
Infectious pancreatic necrosis virus (strain Jasper) (IPNV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Birnaviridae; Aquabirnavirus; Infectious pancreatic necrosis virus
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
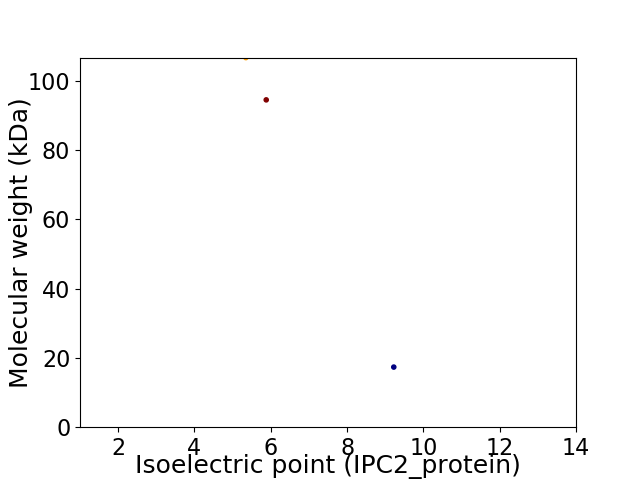
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P22173|RDRP_IPNVJ RNA-directed RNA polymerase OS=Infectious pancreatic necrosis virus (strain Jasper) OX=11003 GN=VP1 PE=1 SV=1
MM1 pKa = 6.85STSKK5 pKa = 9.92ATATYY10 pKa = 9.68LRR12 pKa = 11.84SIMLPEE18 pKa = 4.78NGPASIPDD26 pKa = 5.04DD27 pKa = 3.12ITEE30 pKa = 3.99RR31 pKa = 11.84HH32 pKa = 5.89ILKK35 pKa = 10.07QEE37 pKa = 3.84TSSYY41 pKa = 9.6NLEE44 pKa = 4.05VSEE47 pKa = 4.7SGSGLLVCFPGAPGSRR63 pKa = 11.84VGAHH67 pKa = 5.62YY68 pKa = 10.33RR69 pKa = 11.84WNLNQTALEE78 pKa = 3.92FDD80 pKa = 3.2QWLEE84 pKa = 3.86TSQDD88 pKa = 3.07LKK90 pKa = 11.21KK91 pKa = 10.75AFNYY95 pKa = 9.81GRR97 pKa = 11.84LISRR101 pKa = 11.84KK102 pKa = 9.31YY103 pKa = 10.36DD104 pKa = 3.38IQSSTLPAGLYY115 pKa = 10.34ALNGTLNAATFEE127 pKa = 4.46GSLSEE132 pKa = 4.62VEE134 pKa = 4.23SLTYY138 pKa = 10.53NSLMSLTTNPQDD150 pKa = 3.31KK151 pKa = 10.9VNNQLVTKK159 pKa = 10.33GITVLNLPTGFDD171 pKa = 3.17KK172 pKa = 10.97PYY174 pKa = 10.8VRR176 pKa = 11.84LEE178 pKa = 4.3DD179 pKa = 3.78EE180 pKa = 4.8TPQGPQSMNGARR192 pKa = 11.84MRR194 pKa = 11.84CTAAIAPRR202 pKa = 11.84RR203 pKa = 11.84YY204 pKa = 10.22EE205 pKa = 3.45IDD207 pKa = 3.67LPSEE211 pKa = 4.08RR212 pKa = 11.84LPTVAATGTPTTIYY226 pKa = 10.0EE227 pKa = 4.25GNADD231 pKa = 3.82IVNSTAVTGDD241 pKa = 2.71ITFQLEE247 pKa = 4.12AEE249 pKa = 4.54PVNEE253 pKa = 3.99TRR255 pKa = 11.84FDD257 pKa = 4.63FILQFLGLDD266 pKa = 3.17NDD268 pKa = 4.13VPVVTVTSSTLVTADD283 pKa = 3.27NYY285 pKa = 10.84RR286 pKa = 11.84GASAKK291 pKa = 8.3FTQSIPTEE299 pKa = 4.46MITKK303 pKa = 9.77PITRR307 pKa = 11.84VKK309 pKa = 10.35LAYY312 pKa = 9.8QLNQQTAIANAATLGAKK329 pKa = 10.23GPASVSFSSGNGNVPGVLRR348 pKa = 11.84PITLVAYY355 pKa = 9.89EE356 pKa = 4.46KK357 pKa = 7.78MTPQSILTVAGVSNYY372 pKa = 9.47EE373 pKa = 4.59LIPNPDD379 pKa = 3.58LLKK382 pKa = 11.45NMVTKK387 pKa = 10.51YY388 pKa = 10.85GKK390 pKa = 10.23YY391 pKa = 10.23DD392 pKa = 3.7PEE394 pKa = 4.22GLNYY398 pKa = 10.95AKK400 pKa = 10.06MILSHH405 pKa = 7.46RR406 pKa = 11.84EE407 pKa = 3.56EE408 pKa = 4.78LDD410 pKa = 2.89IRR412 pKa = 11.84TVWRR416 pKa = 11.84TEE418 pKa = 3.79EE419 pKa = 3.97YY420 pKa = 10.52KK421 pKa = 10.87EE422 pKa = 3.62RR423 pKa = 11.84TRR425 pKa = 11.84AFKK428 pKa = 10.48EE429 pKa = 3.83ITDD432 pKa = 4.1FTSDD436 pKa = 4.33LPTSKK441 pKa = 10.55AWGWRR446 pKa = 11.84DD447 pKa = 3.21LVRR450 pKa = 11.84GIRR453 pKa = 11.84KK454 pKa = 8.6VAAPVLSTLFPMAAPLIGAADD475 pKa = 3.69QFIGDD480 pKa = 3.86LTKK483 pKa = 10.61TNSAGGRR490 pKa = 11.84YY491 pKa = 9.11LSHH494 pKa = 6.97AAGGRR499 pKa = 11.84YY500 pKa = 8.91HH501 pKa = 7.51DD502 pKa = 5.76VMDD505 pKa = 3.94SWASGSEE512 pKa = 3.51AGSYY516 pKa = 10.26SKK518 pKa = 10.62HH519 pKa = 5.68LKK521 pKa = 8.54TRR523 pKa = 11.84LEE525 pKa = 4.22SNNYY529 pKa = 9.89EE530 pKa = 4.21EE531 pKa = 5.46VEE533 pKa = 4.19LPKK536 pKa = 9.21PTKK539 pKa = 10.28GVIFPVVHH547 pKa = 5.68TVEE550 pKa = 4.43SAPGEE555 pKa = 4.18AFGSLVVVIPEE566 pKa = 5.11AYY568 pKa = 9.5PEE570 pKa = 4.38LLDD573 pKa = 4.73PNQQVLSYY581 pKa = 9.38FKK583 pKa = 11.03NDD585 pKa = 3.51TGCVWGIGEE594 pKa = 5.44DD595 pKa = 3.57IPFEE599 pKa = 5.1GDD601 pKa = 3.46DD602 pKa = 3.54MCYY605 pKa = 9.28TALPLKK611 pKa = 9.86EE612 pKa = 3.89IKK614 pKa = 10.42RR615 pKa = 11.84NGNIVVEE622 pKa = 4.81KK623 pKa = 10.34IFAGPAMGPSSQLALSLLVNDD644 pKa = 4.18IDD646 pKa = 3.83EE647 pKa = 5.54GIPRR651 pKa = 11.84MVFTGEE657 pKa = 3.79IADD660 pKa = 4.48DD661 pKa = 4.32EE662 pKa = 4.63EE663 pKa = 4.8TVIPICGVDD672 pKa = 3.34IKK674 pKa = 11.15AIAAHH679 pKa = 5.38EE680 pKa = 4.63HH681 pKa = 5.69GLPLIGCQPGVDD693 pKa = 3.11EE694 pKa = 4.62MVANTSLASHH704 pKa = 7.66LIQGGALPVQKK715 pKa = 10.73AQGACRR721 pKa = 11.84RR722 pKa = 11.84IKK724 pKa = 10.9YY725 pKa = 9.71LGQLMRR731 pKa = 11.84TTASGMDD738 pKa = 3.61AEE740 pKa = 4.72LQGLLQATMARR751 pKa = 11.84AKK753 pKa = 9.47EE754 pKa = 4.15VKK756 pKa = 9.93DD757 pKa = 3.6AEE759 pKa = 4.44VFKK762 pKa = 10.84LLKK765 pKa = 10.54LMSWTRR771 pKa = 11.84KK772 pKa = 9.62NDD774 pKa = 3.71LTDD777 pKa = 5.05HH778 pKa = 5.96MYY780 pKa = 10.29EE781 pKa = 3.96WSKK784 pKa = 11.28EE785 pKa = 3.95DD786 pKa = 3.38PDD788 pKa = 4.52AIKK791 pKa = 10.46FGRR794 pKa = 11.84LVSTPPKK801 pKa = 9.86HH802 pKa = 5.49QEE804 pKa = 3.87KK805 pKa = 10.39PKK807 pKa = 11.16GPDD810 pKa = 2.69QHH812 pKa = 6.27TAQEE816 pKa = 4.12AKK818 pKa = 9.17ATRR821 pKa = 11.84ISLDD825 pKa = 3.29AVKK828 pKa = 10.64AGADD832 pKa = 3.54FASPEE837 pKa = 4.06WIAEE841 pKa = 3.88NNYY844 pKa = 10.01RR845 pKa = 11.84GPSPGQFKK853 pKa = 11.0YY854 pKa = 11.47YY855 pKa = 9.51MITGRR860 pKa = 11.84VPNPGEE866 pKa = 3.84EE867 pKa = 4.23YY868 pKa = 9.96EE869 pKa = 4.56DD870 pKa = 4.02YY871 pKa = 10.81VRR873 pKa = 11.84KK874 pKa = 9.51PITRR878 pKa = 11.84PTDD881 pKa = 2.99MDD883 pKa = 4.24KK884 pKa = 10.54IRR886 pKa = 11.84RR887 pKa = 11.84LANSVYY893 pKa = 10.06GLPHH897 pKa = 6.31QEE899 pKa = 3.81PAPDD903 pKa = 3.89DD904 pKa = 4.21FYY906 pKa = 11.43QAVVEE911 pKa = 4.34VFAEE915 pKa = 4.05NGGRR919 pKa = 11.84GPDD922 pKa = 3.4QDD924 pKa = 3.5QMQDD928 pKa = 3.21LRR930 pKa = 11.84DD931 pKa = 3.54LARR934 pKa = 11.84QMKK937 pKa = 9.58RR938 pKa = 11.84RR939 pKa = 11.84PRR941 pKa = 11.84PAEE944 pKa = 3.49TRR946 pKa = 11.84RR947 pKa = 11.84QTKK950 pKa = 8.26TPPRR954 pKa = 11.84AATSSGSRR962 pKa = 11.84FTPSGDD968 pKa = 3.49DD969 pKa = 3.82GEE971 pKa = 4.6VV972 pKa = 2.82
MM1 pKa = 6.85STSKK5 pKa = 9.92ATATYY10 pKa = 9.68LRR12 pKa = 11.84SIMLPEE18 pKa = 4.78NGPASIPDD26 pKa = 5.04DD27 pKa = 3.12ITEE30 pKa = 3.99RR31 pKa = 11.84HH32 pKa = 5.89ILKK35 pKa = 10.07QEE37 pKa = 3.84TSSYY41 pKa = 9.6NLEE44 pKa = 4.05VSEE47 pKa = 4.7SGSGLLVCFPGAPGSRR63 pKa = 11.84VGAHH67 pKa = 5.62YY68 pKa = 10.33RR69 pKa = 11.84WNLNQTALEE78 pKa = 3.92FDD80 pKa = 3.2QWLEE84 pKa = 3.86TSQDD88 pKa = 3.07LKK90 pKa = 11.21KK91 pKa = 10.75AFNYY95 pKa = 9.81GRR97 pKa = 11.84LISRR101 pKa = 11.84KK102 pKa = 9.31YY103 pKa = 10.36DD104 pKa = 3.38IQSSTLPAGLYY115 pKa = 10.34ALNGTLNAATFEE127 pKa = 4.46GSLSEE132 pKa = 4.62VEE134 pKa = 4.23SLTYY138 pKa = 10.53NSLMSLTTNPQDD150 pKa = 3.31KK151 pKa = 10.9VNNQLVTKK159 pKa = 10.33GITVLNLPTGFDD171 pKa = 3.17KK172 pKa = 10.97PYY174 pKa = 10.8VRR176 pKa = 11.84LEE178 pKa = 4.3DD179 pKa = 3.78EE180 pKa = 4.8TPQGPQSMNGARR192 pKa = 11.84MRR194 pKa = 11.84CTAAIAPRR202 pKa = 11.84RR203 pKa = 11.84YY204 pKa = 10.22EE205 pKa = 3.45IDD207 pKa = 3.67LPSEE211 pKa = 4.08RR212 pKa = 11.84LPTVAATGTPTTIYY226 pKa = 10.0EE227 pKa = 4.25GNADD231 pKa = 3.82IVNSTAVTGDD241 pKa = 2.71ITFQLEE247 pKa = 4.12AEE249 pKa = 4.54PVNEE253 pKa = 3.99TRR255 pKa = 11.84FDD257 pKa = 4.63FILQFLGLDD266 pKa = 3.17NDD268 pKa = 4.13VPVVTVTSSTLVTADD283 pKa = 3.27NYY285 pKa = 10.84RR286 pKa = 11.84GASAKK291 pKa = 8.3FTQSIPTEE299 pKa = 4.46MITKK303 pKa = 9.77PITRR307 pKa = 11.84VKK309 pKa = 10.35LAYY312 pKa = 9.8QLNQQTAIANAATLGAKK329 pKa = 10.23GPASVSFSSGNGNVPGVLRR348 pKa = 11.84PITLVAYY355 pKa = 9.89EE356 pKa = 4.46KK357 pKa = 7.78MTPQSILTVAGVSNYY372 pKa = 9.47EE373 pKa = 4.59LIPNPDD379 pKa = 3.58LLKK382 pKa = 11.45NMVTKK387 pKa = 10.51YY388 pKa = 10.85GKK390 pKa = 10.23YY391 pKa = 10.23DD392 pKa = 3.7PEE394 pKa = 4.22GLNYY398 pKa = 10.95AKK400 pKa = 10.06MILSHH405 pKa = 7.46RR406 pKa = 11.84EE407 pKa = 3.56EE408 pKa = 4.78LDD410 pKa = 2.89IRR412 pKa = 11.84TVWRR416 pKa = 11.84TEE418 pKa = 3.79EE419 pKa = 3.97YY420 pKa = 10.52KK421 pKa = 10.87EE422 pKa = 3.62RR423 pKa = 11.84TRR425 pKa = 11.84AFKK428 pKa = 10.48EE429 pKa = 3.83ITDD432 pKa = 4.1FTSDD436 pKa = 4.33LPTSKK441 pKa = 10.55AWGWRR446 pKa = 11.84DD447 pKa = 3.21LVRR450 pKa = 11.84GIRR453 pKa = 11.84KK454 pKa = 8.6VAAPVLSTLFPMAAPLIGAADD475 pKa = 3.69QFIGDD480 pKa = 3.86LTKK483 pKa = 10.61TNSAGGRR490 pKa = 11.84YY491 pKa = 9.11LSHH494 pKa = 6.97AAGGRR499 pKa = 11.84YY500 pKa = 8.91HH501 pKa = 7.51DD502 pKa = 5.76VMDD505 pKa = 3.94SWASGSEE512 pKa = 3.51AGSYY516 pKa = 10.26SKK518 pKa = 10.62HH519 pKa = 5.68LKK521 pKa = 8.54TRR523 pKa = 11.84LEE525 pKa = 4.22SNNYY529 pKa = 9.89EE530 pKa = 4.21EE531 pKa = 5.46VEE533 pKa = 4.19LPKK536 pKa = 9.21PTKK539 pKa = 10.28GVIFPVVHH547 pKa = 5.68TVEE550 pKa = 4.43SAPGEE555 pKa = 4.18AFGSLVVVIPEE566 pKa = 5.11AYY568 pKa = 9.5PEE570 pKa = 4.38LLDD573 pKa = 4.73PNQQVLSYY581 pKa = 9.38FKK583 pKa = 11.03NDD585 pKa = 3.51TGCVWGIGEE594 pKa = 5.44DD595 pKa = 3.57IPFEE599 pKa = 5.1GDD601 pKa = 3.46DD602 pKa = 3.54MCYY605 pKa = 9.28TALPLKK611 pKa = 9.86EE612 pKa = 3.89IKK614 pKa = 10.42RR615 pKa = 11.84NGNIVVEE622 pKa = 4.81KK623 pKa = 10.34IFAGPAMGPSSQLALSLLVNDD644 pKa = 4.18IDD646 pKa = 3.83EE647 pKa = 5.54GIPRR651 pKa = 11.84MVFTGEE657 pKa = 3.79IADD660 pKa = 4.48DD661 pKa = 4.32EE662 pKa = 4.63EE663 pKa = 4.8TVIPICGVDD672 pKa = 3.34IKK674 pKa = 11.15AIAAHH679 pKa = 5.38EE680 pKa = 4.63HH681 pKa = 5.69GLPLIGCQPGVDD693 pKa = 3.11EE694 pKa = 4.62MVANTSLASHH704 pKa = 7.66LIQGGALPVQKK715 pKa = 10.73AQGACRR721 pKa = 11.84RR722 pKa = 11.84IKK724 pKa = 10.9YY725 pKa = 9.71LGQLMRR731 pKa = 11.84TTASGMDD738 pKa = 3.61AEE740 pKa = 4.72LQGLLQATMARR751 pKa = 11.84AKK753 pKa = 9.47EE754 pKa = 4.15VKK756 pKa = 9.93DD757 pKa = 3.6AEE759 pKa = 4.44VFKK762 pKa = 10.84LLKK765 pKa = 10.54LMSWTRR771 pKa = 11.84KK772 pKa = 9.62NDD774 pKa = 3.71LTDD777 pKa = 5.05HH778 pKa = 5.96MYY780 pKa = 10.29EE781 pKa = 3.96WSKK784 pKa = 11.28EE785 pKa = 3.95DD786 pKa = 3.38PDD788 pKa = 4.52AIKK791 pKa = 10.46FGRR794 pKa = 11.84LVSTPPKK801 pKa = 9.86HH802 pKa = 5.49QEE804 pKa = 3.87KK805 pKa = 10.39PKK807 pKa = 11.16GPDD810 pKa = 2.69QHH812 pKa = 6.27TAQEE816 pKa = 4.12AKK818 pKa = 9.17ATRR821 pKa = 11.84ISLDD825 pKa = 3.29AVKK828 pKa = 10.64AGADD832 pKa = 3.54FASPEE837 pKa = 4.06WIAEE841 pKa = 3.88NNYY844 pKa = 10.01RR845 pKa = 11.84GPSPGQFKK853 pKa = 11.0YY854 pKa = 11.47YY855 pKa = 9.51MITGRR860 pKa = 11.84VPNPGEE866 pKa = 3.84EE867 pKa = 4.23YY868 pKa = 9.96EE869 pKa = 4.56DD870 pKa = 4.02YY871 pKa = 10.81VRR873 pKa = 11.84KK874 pKa = 9.51PITRR878 pKa = 11.84PTDD881 pKa = 2.99MDD883 pKa = 4.24KK884 pKa = 10.54IRR886 pKa = 11.84RR887 pKa = 11.84LANSVYY893 pKa = 10.06GLPHH897 pKa = 6.31QEE899 pKa = 3.81PAPDD903 pKa = 3.89DD904 pKa = 4.21FYY906 pKa = 11.43QAVVEE911 pKa = 4.34VFAEE915 pKa = 4.05NGGRR919 pKa = 11.84GPDD922 pKa = 3.4QDD924 pKa = 3.5QMQDD928 pKa = 3.21LRR930 pKa = 11.84DD931 pKa = 3.54LARR934 pKa = 11.84QMKK937 pKa = 9.58RR938 pKa = 11.84RR939 pKa = 11.84PRR941 pKa = 11.84PAEE944 pKa = 3.49TRR946 pKa = 11.84RR947 pKa = 11.84QTKK950 pKa = 8.26TPPRR954 pKa = 11.84AATSSGSRR962 pKa = 11.84FTPSGDD968 pKa = 3.49DD969 pKa = 3.82GEE971 pKa = 4.6VV972 pKa = 2.82
Molecular weight: 106.67 kDa
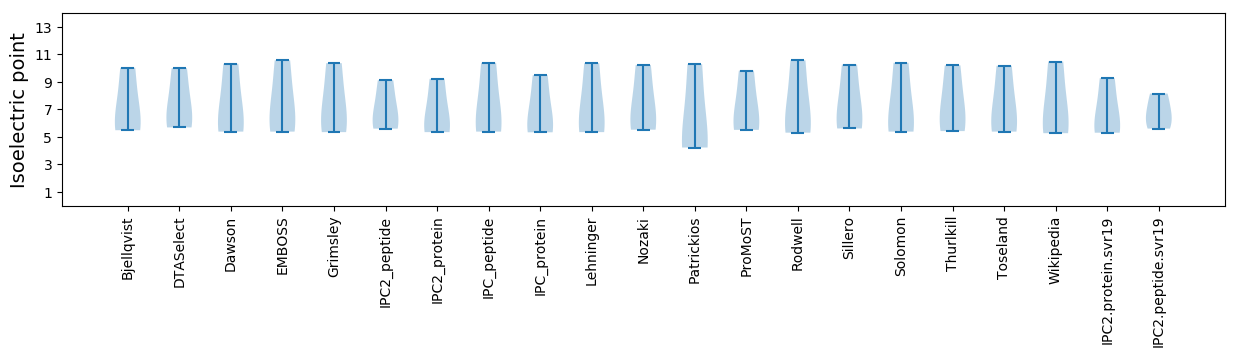
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P22931|VP5_IPNVJ Protein VP5 OS=Infectious pancreatic necrosis virus (strain Jasper) OX=11003 GN=VP5 PE=3 SV=1
MM1 pKa = 7.71AKK3 pKa = 10.29ALSNKK8 pKa = 6.69PTNILIYY15 pKa = 9.45MNHH18 pKa = 5.76EE19 pKa = 4.53HH20 pKa = 6.41IQGNRR25 pKa = 11.84NLLEE29 pKa = 3.32IHH31 pKa = 5.78YY32 pKa = 10.24ASRR35 pKa = 11.84EE36 pKa = 4.02WASKK40 pKa = 10.27HH41 pKa = 5.16SGRR44 pKa = 11.84HH45 pKa = 3.75NRR47 pKa = 11.84EE48 pKa = 3.92AYY50 pKa = 8.14TKK52 pKa = 10.12TRR54 pKa = 11.84DD55 pKa = 3.76LVIQLRR61 pKa = 11.84GIRR64 pKa = 11.84IRR66 pKa = 11.84KK67 pKa = 6.51WASCLLPRR75 pKa = 11.84SSWIQGRR82 pKa = 11.84CPLQVEE88 pKa = 4.5SEE90 pKa = 4.15PDD92 pKa = 3.2GTRR95 pKa = 11.84IRR97 pKa = 11.84PVARR101 pKa = 11.84DD102 pKa = 3.21VTGPKK107 pKa = 10.08EE108 pKa = 4.45GIQLRR113 pKa = 11.84EE114 pKa = 3.66TDD116 pKa = 3.56LTEE119 pKa = 4.03IRR121 pKa = 11.84HH122 pKa = 6.1PEE124 pKa = 4.03LNPSRR129 pKa = 11.84WSVCTQWDD137 pKa = 3.64PEE139 pKa = 4.23RR140 pKa = 11.84CHH142 pKa = 7.07LRR144 pKa = 11.84RR145 pKa = 11.84KK146 pKa = 9.69SVV148 pKa = 2.76
MM1 pKa = 7.71AKK3 pKa = 10.29ALSNKK8 pKa = 6.69PTNILIYY15 pKa = 9.45MNHH18 pKa = 5.76EE19 pKa = 4.53HH20 pKa = 6.41IQGNRR25 pKa = 11.84NLLEE29 pKa = 3.32IHH31 pKa = 5.78YY32 pKa = 10.24ASRR35 pKa = 11.84EE36 pKa = 4.02WASKK40 pKa = 10.27HH41 pKa = 5.16SGRR44 pKa = 11.84HH45 pKa = 3.75NRR47 pKa = 11.84EE48 pKa = 3.92AYY50 pKa = 8.14TKK52 pKa = 10.12TRR54 pKa = 11.84DD55 pKa = 3.76LVIQLRR61 pKa = 11.84GIRR64 pKa = 11.84IRR66 pKa = 11.84KK67 pKa = 6.51WASCLLPRR75 pKa = 11.84SSWIQGRR82 pKa = 11.84CPLQVEE88 pKa = 4.5SEE90 pKa = 4.15PDD92 pKa = 3.2GTRR95 pKa = 11.84IRR97 pKa = 11.84PVARR101 pKa = 11.84DD102 pKa = 3.21VTGPKK107 pKa = 10.08EE108 pKa = 4.45GIQLRR113 pKa = 11.84EE114 pKa = 3.66TDD116 pKa = 3.56LTEE119 pKa = 4.03IRR121 pKa = 11.84HH122 pKa = 6.1PEE124 pKa = 4.03LNPSRR129 pKa = 11.84WSVCTQWDD137 pKa = 3.64PEE139 pKa = 4.23RR140 pKa = 11.84CHH142 pKa = 7.07LRR144 pKa = 11.84RR145 pKa = 11.84KK146 pKa = 9.69SVV148 pKa = 2.76
Molecular weight: 17.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1965 |
148 |
972 |
655.0 |
72.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.99 ± 0.804 | 0.712 ± 0.441 |
5.802 ± 0.541 | 6.463 ± 0.244 |
2.646 ± 0.577 | 6.565 ± 0.697 |
1.527 ± 0.699 | 5.089 ± 0.513 |
5.903 ± 0.624 | 9.415 ± 0.711 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.545 ± 0.278 | 4.682 ± 0.316 |
6.768 ± 0.16 | 3.868 ± 0.054 |
5.7 ± 1.55 | 6.412 ± 0.274 |
7.786 ± 0.561 | 5.344 ± 0.586 |
1.476 ± 0.471 | 3.308 ± 0.288 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
