
Candidatus Nitrosocaldus cavascurensis
Taxonomy: cellular organisms; Archaea; TACK group; Thaumarchaeota; Nitrososphaeria; Candidatus Nitrosocaldales; Candidatus Nitrosocaldaceae; Candidatus Nitrosocaldus
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
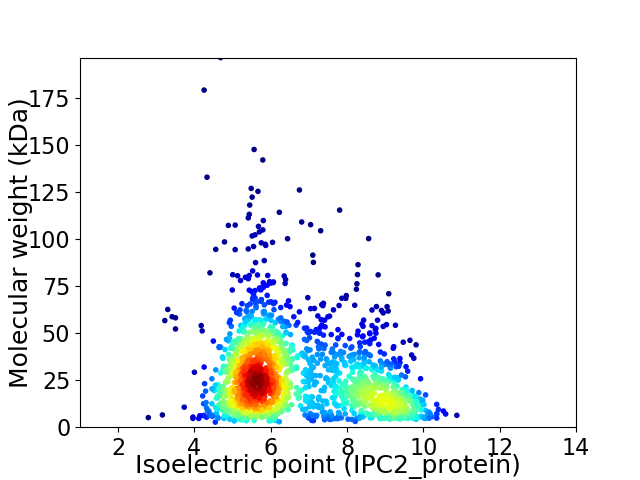
Virtual 2D-PAGE plot for 1766 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2K5ARG9|A0A2K5ARG9_9ARCH Uncharacterized protein OS=Candidatus Nitrosocaldus cavascurensis OX=2058097 GN=NCAV_1022 PE=4 SV=1
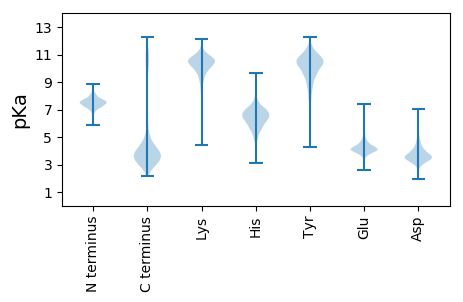
MM1 pKa = 7.78AEE3 pKa = 3.87NGGITRR9 pKa = 11.84AEE11 pKa = 4.01KK12 pKa = 10.42RR13 pKa = 11.84VTSATAITITITTILLLMVVSMTVTDD39 pKa = 5.21GIRR42 pKa = 11.84TVYY45 pKa = 10.18AQSATITAEE54 pKa = 4.47CNGHH58 pKa = 5.0TIEE61 pKa = 5.45RR62 pKa = 11.84GNFDD66 pKa = 4.19ADD68 pKa = 3.27GDD70 pKa = 4.36DD71 pKa = 3.69EE72 pKa = 5.05VKK74 pKa = 10.83VDD76 pKa = 3.41GTVYY80 pKa = 11.06NDD82 pKa = 3.56GDD84 pKa = 4.65TITLPSGTYY93 pKa = 8.81TIRR96 pKa = 11.84IATPSWPTPYY106 pKa = 10.76NGTDD110 pKa = 3.37DD111 pKa = 3.87NDD113 pKa = 4.78FIVGTSGDD121 pKa = 3.71DD122 pKa = 3.6YY123 pKa = 11.33IFGRR127 pKa = 11.84GEE129 pKa = 4.01DD130 pKa = 3.7DD131 pKa = 4.47FIVGFGGNDD140 pKa = 3.38YY141 pKa = 11.09LYY143 pKa = 10.95GDD145 pKa = 4.08VVKK148 pKa = 9.88GTDD151 pKa = 3.72GADD154 pKa = 3.36TLEE157 pKa = 4.71GGDD160 pKa = 6.15DD161 pKa = 3.67ILCGNDD167 pKa = 3.23GDD169 pKa = 4.66DD170 pKa = 3.75YY171 pKa = 11.58LYY173 pKa = 11.4GDD175 pKa = 4.61VIGGGNGNDD184 pKa = 3.32TMTGGNDD191 pKa = 3.1TLYY194 pKa = 11.01GGKK197 pKa = 10.79GNDD200 pKa = 3.3NLFGDD205 pKa = 5.46GIGGAYY211 pKa = 10.15GDD213 pKa = 4.26DD214 pKa = 3.83TLTGGDD220 pKa = 3.44DD221 pKa = 3.32TLYY224 pKa = 11.06GGEE227 pKa = 4.16GDD229 pKa = 4.71DD230 pKa = 3.67WLEE233 pKa = 3.98GDD235 pKa = 4.67VIGGGDD241 pKa = 4.28GDD243 pKa = 3.71NTMTGGNDD251 pKa = 3.07TLYY254 pKa = 10.85GGKK257 pKa = 10.51GSDD260 pKa = 2.98RR261 pKa = 11.84LYY263 pKa = 11.46GDD265 pKa = 5.19GIWGEE270 pKa = 4.59DD271 pKa = 3.74GDD273 pKa = 4.15NTMTGGNDD281 pKa = 3.07TLYY284 pKa = 10.85GGKK287 pKa = 10.51GSDD290 pKa = 2.98RR291 pKa = 11.84LYY293 pKa = 11.27GDD295 pKa = 4.82GIGGGVGNDD304 pKa = 3.45TLTGGDD310 pKa = 3.81DD311 pKa = 3.63TLKK314 pKa = 11.24GGDD317 pKa = 4.05GDD319 pKa = 4.38DD320 pKa = 3.8YY321 pKa = 11.61LYY323 pKa = 11.26GDD325 pKa = 5.18SIDD328 pKa = 4.47GGPGVDD334 pKa = 5.11KK335 pKa = 10.96IQGGNDD341 pKa = 3.06TLYY344 pKa = 11.01GGKK347 pKa = 10.74GNDD350 pKa = 3.41KK351 pKa = 10.98LFGDD355 pKa = 5.47GIWGDD360 pKa = 3.72ADD362 pKa = 3.94NDD364 pKa = 3.95TAYY367 pKa = 10.69GGNDD371 pKa = 3.35VIDD374 pKa = 4.03ARR376 pKa = 11.84DD377 pKa = 3.6NVEE380 pKa = 5.08DD381 pKa = 4.13NDD383 pKa = 5.44SIDD386 pKa = 3.25GDD388 pKa = 4.3YY389 pKa = 10.75IILAEE394 pKa = 4.29STTEE398 pKa = 4.07GNQDD402 pKa = 2.62ICASDD407 pKa = 4.02PEE409 pKa = 4.33DD410 pKa = 3.63TEE412 pKa = 4.95VNCEE416 pKa = 3.81YY417 pKa = 11.06HH418 pKa = 7.41DD419 pKa = 4.01ISGLATLTTDD429 pKa = 3.62SATIPLGGSVQIIFNSSVDD448 pKa = 3.75NPVKK452 pKa = 9.74ITNLRR457 pKa = 11.84VITPTGNTCTYY468 pKa = 9.78TGTLPIIVPSNPFTATYY485 pKa = 9.22PDD487 pKa = 3.72NFIDD491 pKa = 4.18IDD493 pKa = 3.79TGTDD497 pKa = 3.38CDD499 pKa = 3.73TSAVGEE505 pKa = 4.28YY506 pKa = 9.81TVVAEE511 pKa = 4.63TEE513 pKa = 4.06VGDD516 pKa = 5.26PITTNFNISFNVVPEE531 pKa = 4.11AVVGAVGMVGTGFLSILLYY550 pKa = 10.4RR551 pKa = 11.84RR552 pKa = 11.84LRR554 pKa = 11.84GKK556 pKa = 9.14SRR558 pKa = 11.84KK559 pKa = 7.93EE560 pKa = 3.55
MM1 pKa = 7.78AEE3 pKa = 3.87NGGITRR9 pKa = 11.84AEE11 pKa = 4.01KK12 pKa = 10.42RR13 pKa = 11.84VTSATAITITITTILLLMVVSMTVTDD39 pKa = 5.21GIRR42 pKa = 11.84TVYY45 pKa = 10.18AQSATITAEE54 pKa = 4.47CNGHH58 pKa = 5.0TIEE61 pKa = 5.45RR62 pKa = 11.84GNFDD66 pKa = 4.19ADD68 pKa = 3.27GDD70 pKa = 4.36DD71 pKa = 3.69EE72 pKa = 5.05VKK74 pKa = 10.83VDD76 pKa = 3.41GTVYY80 pKa = 11.06NDD82 pKa = 3.56GDD84 pKa = 4.65TITLPSGTYY93 pKa = 8.81TIRR96 pKa = 11.84IATPSWPTPYY106 pKa = 10.76NGTDD110 pKa = 3.37DD111 pKa = 3.87NDD113 pKa = 4.78FIVGTSGDD121 pKa = 3.71DD122 pKa = 3.6YY123 pKa = 11.33IFGRR127 pKa = 11.84GEE129 pKa = 4.01DD130 pKa = 3.7DD131 pKa = 4.47FIVGFGGNDD140 pKa = 3.38YY141 pKa = 11.09LYY143 pKa = 10.95GDD145 pKa = 4.08VVKK148 pKa = 9.88GTDD151 pKa = 3.72GADD154 pKa = 3.36TLEE157 pKa = 4.71GGDD160 pKa = 6.15DD161 pKa = 3.67ILCGNDD167 pKa = 3.23GDD169 pKa = 4.66DD170 pKa = 3.75YY171 pKa = 11.58LYY173 pKa = 11.4GDD175 pKa = 4.61VIGGGNGNDD184 pKa = 3.32TMTGGNDD191 pKa = 3.1TLYY194 pKa = 11.01GGKK197 pKa = 10.79GNDD200 pKa = 3.3NLFGDD205 pKa = 5.46GIGGAYY211 pKa = 10.15GDD213 pKa = 4.26DD214 pKa = 3.83TLTGGDD220 pKa = 3.44DD221 pKa = 3.32TLYY224 pKa = 11.06GGEE227 pKa = 4.16GDD229 pKa = 4.71DD230 pKa = 3.67WLEE233 pKa = 3.98GDD235 pKa = 4.67VIGGGDD241 pKa = 4.28GDD243 pKa = 3.71NTMTGGNDD251 pKa = 3.07TLYY254 pKa = 10.85GGKK257 pKa = 10.51GSDD260 pKa = 2.98RR261 pKa = 11.84LYY263 pKa = 11.46GDD265 pKa = 5.19GIWGEE270 pKa = 4.59DD271 pKa = 3.74GDD273 pKa = 4.15NTMTGGNDD281 pKa = 3.07TLYY284 pKa = 10.85GGKK287 pKa = 10.51GSDD290 pKa = 2.98RR291 pKa = 11.84LYY293 pKa = 11.27GDD295 pKa = 4.82GIGGGVGNDD304 pKa = 3.45TLTGGDD310 pKa = 3.81DD311 pKa = 3.63TLKK314 pKa = 11.24GGDD317 pKa = 4.05GDD319 pKa = 4.38DD320 pKa = 3.8YY321 pKa = 11.61LYY323 pKa = 11.26GDD325 pKa = 5.18SIDD328 pKa = 4.47GGPGVDD334 pKa = 5.11KK335 pKa = 10.96IQGGNDD341 pKa = 3.06TLYY344 pKa = 11.01GGKK347 pKa = 10.74GNDD350 pKa = 3.41KK351 pKa = 10.98LFGDD355 pKa = 5.47GIWGDD360 pKa = 3.72ADD362 pKa = 3.94NDD364 pKa = 3.95TAYY367 pKa = 10.69GGNDD371 pKa = 3.35VIDD374 pKa = 4.03ARR376 pKa = 11.84DD377 pKa = 3.6NVEE380 pKa = 5.08DD381 pKa = 4.13NDD383 pKa = 5.44SIDD386 pKa = 3.25GDD388 pKa = 4.3YY389 pKa = 10.75IILAEE394 pKa = 4.29STTEE398 pKa = 4.07GNQDD402 pKa = 2.62ICASDD407 pKa = 4.02PEE409 pKa = 4.33DD410 pKa = 3.63TEE412 pKa = 4.95VNCEE416 pKa = 3.81YY417 pKa = 11.06HH418 pKa = 7.41DD419 pKa = 4.01ISGLATLTTDD429 pKa = 3.62SATIPLGGSVQIIFNSSVDD448 pKa = 3.75NPVKK452 pKa = 9.74ITNLRR457 pKa = 11.84VITPTGNTCTYY468 pKa = 9.78TGTLPIIVPSNPFTATYY485 pKa = 9.22PDD487 pKa = 3.72NFIDD491 pKa = 4.18IDD493 pKa = 3.79TGTDD497 pKa = 3.38CDD499 pKa = 3.73TSAVGEE505 pKa = 4.28YY506 pKa = 9.81TVVAEE511 pKa = 4.63TEE513 pKa = 4.06VGDD516 pKa = 5.26PITTNFNISFNVVPEE531 pKa = 4.11AVVGAVGMVGTGFLSILLYY550 pKa = 10.4RR551 pKa = 11.84RR552 pKa = 11.84LRR554 pKa = 11.84GKK556 pKa = 9.14SRR558 pKa = 11.84KK559 pKa = 7.93EE560 pKa = 3.55
Molecular weight: 58.09 kDa
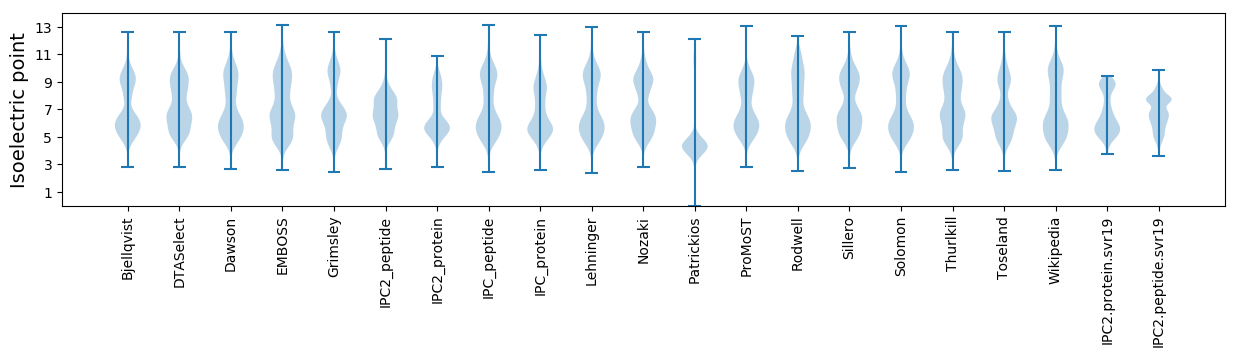
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2K5ASG8|A0A2K5ASG8_9ARCH Uncharacterized protein OS=Candidatus Nitrosocaldus cavascurensis OX=2058097 GN=NCAV_1432 PE=4 SV=1
MM1 pKa = 7.81GSRR4 pKa = 11.84KK5 pKa = 7.97TKK7 pKa = 8.99PRR9 pKa = 11.84KK10 pKa = 8.81NRR12 pKa = 11.84LMKK15 pKa = 9.39KK16 pKa = 7.91TKK18 pKa = 10.09QNSPVPLWVILKK30 pKa = 7.0TNRR33 pKa = 11.84RR34 pKa = 11.84VRR36 pKa = 11.84RR37 pKa = 11.84NPQQRR42 pKa = 11.84HH43 pKa = 3.83WRR45 pKa = 11.84RR46 pKa = 11.84SSVDD50 pKa = 3.09VGG52 pKa = 3.49
MM1 pKa = 7.81GSRR4 pKa = 11.84KK5 pKa = 7.97TKK7 pKa = 8.99PRR9 pKa = 11.84KK10 pKa = 8.81NRR12 pKa = 11.84LMKK15 pKa = 9.39KK16 pKa = 7.91TKK18 pKa = 10.09QNSPVPLWVILKK30 pKa = 7.0TNRR33 pKa = 11.84RR34 pKa = 11.84VRR36 pKa = 11.84RR37 pKa = 11.84NPQQRR42 pKa = 11.84HH43 pKa = 3.83WRR45 pKa = 11.84RR46 pKa = 11.84SSVDD50 pKa = 3.09VGG52 pKa = 3.49
Molecular weight: 6.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
461185 |
24 |
1803 |
261.1 |
29.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.613 ± 0.059 | 1.06 ± 0.024 |
6.09 ± 0.061 | 6.57 ± 0.074 |
3.18 ± 0.04 | 6.909 ± 0.064 |
1.976 ± 0.025 | 8.286 ± 0.052 |
5.683 ± 0.058 | 9.21 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.435 ± 0.036 | 4.111 ± 0.044 |
3.365 ± 0.04 | 1.994 ± 0.034 |
6.462 ± 0.061 | 7.021 ± 0.069 |
4.302 ± 0.057 | 7.809 ± 0.052 |
0.884 ± 0.021 | 4.04 ± 0.042 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
