
Candida auris (Yeast)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Metschnikowiaceae;
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
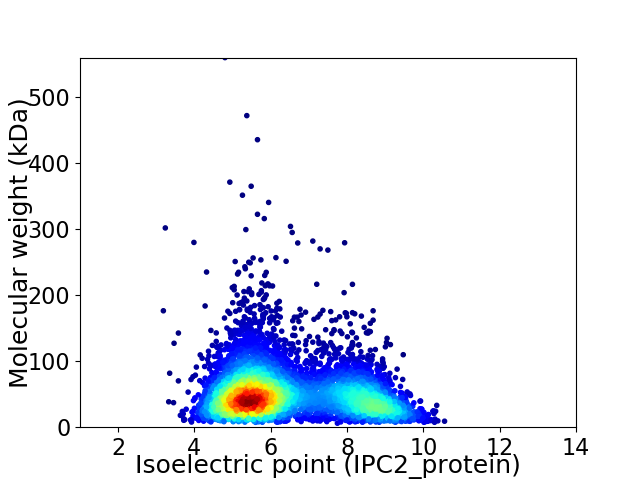
Virtual 2D-PAGE plot for 5409 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H0ZR33|A0A2H0ZR33_CANAR Uncharacterized protein OS=Candida auris OX=498019 GN=B9J08_002729 PE=4 SV=1
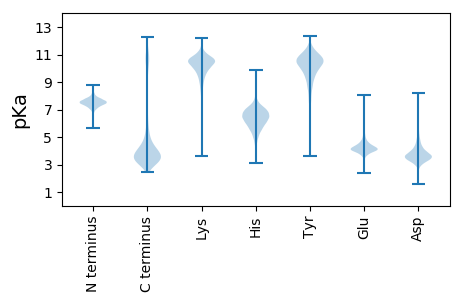
MM1 pKa = 7.57KK2 pKa = 10.38LLSTVATAALLLQKK16 pKa = 10.49ARR18 pKa = 11.84SLTITEE24 pKa = 4.23DD25 pKa = 3.51TVLVSPVNLEE35 pKa = 3.67IGEE38 pKa = 4.44LNINPGVYY46 pKa = 10.31FSIVNNVLTVLGGNLNNDD64 pKa = 3.23GAFYY68 pKa = 9.77VTSTNGLAASVTIASGSIINRR89 pKa = 11.84GDD91 pKa = 3.29LAFNSLKK98 pKa = 11.12ANVITNFNLDD108 pKa = 3.45SVGTFSNTGNMWLGVPIFSAVPPIILGSALDD139 pKa = 3.87WEE141 pKa = 4.77NKK143 pKa = 8.73GMIYY147 pKa = 10.4LRR149 pKa = 11.84QEE151 pKa = 3.94LGGASPITISQVLGAIDD168 pKa = 3.82NSGTICIEE176 pKa = 3.84RR177 pKa = 11.84LNWLQTTTINGAGCVNVQADD197 pKa = 3.72GHH199 pKa = 6.3LQLQISPWSVGEE211 pKa = 4.0DD212 pKa = 3.09QTIYY216 pKa = 11.12LSTPTSALSVLGLEE230 pKa = 4.48PSLLGTKK237 pKa = 9.05TYY239 pKa = 11.13NVVGFGGGNTIGINLGFTSYY259 pKa = 10.56SYY261 pKa = 11.12SGSTLTLSFFLGVFKK276 pKa = 10.9INFNIGEE283 pKa = 4.58GYY285 pKa = 10.06SADD288 pKa = 3.37GFSTNGPGNSGTQITYY304 pKa = 10.21DD305 pKa = 3.37GPYY308 pKa = 9.31PGSVPDD314 pKa = 3.87KK315 pKa = 10.07CLCKK319 pKa = 10.57DD320 pKa = 3.9FPTPPTEE327 pKa = 4.35PSSSSSSSSSAPPSSSAPEE346 pKa = 3.86SSSTSEE352 pKa = 4.09SSSSEE357 pKa = 3.92SSSSSEE363 pKa = 4.02PPSSEE368 pKa = 4.07TSSSEE373 pKa = 4.15PPSSSQPPSSSEE385 pKa = 3.94PPSSSEE391 pKa = 4.42PPSSSQPPSSSQPPSSSEE409 pKa = 3.94PPSSSEE415 pKa = 4.42PPSSSQPPSSSEE427 pKa = 4.05PPSSSGPSEE436 pKa = 4.19SSGSSSGPSEE446 pKa = 4.2TSGSSSGPSEE456 pKa = 4.17TSGSSTGPSEE466 pKa = 3.93TSGPSEE472 pKa = 4.36SSTGPGNGGEE482 pKa = 4.23TSGPGNGGEE491 pKa = 4.28TSTGPGNGGQPTSGPGNGGEE511 pKa = 4.08PTGPGNGGEE520 pKa = 4.11PTGPGNGGEE529 pKa = 4.11PTGPGNGGQPTGPGNGGEE547 pKa = 4.11PTGPGNGGQPTGPGNGGQPTGPGNGGQPTGPGNGGQPTNGPGNGGQPTGPGGPSEE602 pKa = 4.09TAPGGGNGGGNGNGGGNGNGGGNGNGGGNGNGGEE636 pKa = 4.08NGNGGGNGNGGGNGNGGEE654 pKa = 4.07NGNGGEE660 pKa = 4.17NGNGGGNGTGGEE672 pKa = 4.07NGNGGGNGNGGGNGSGNGSGNGSGNGSGNGSGNGSGNGNGSGNGIEE718 pKa = 4.58TKK720 pKa = 10.03TSQSPTGGAGGPTVSSYY737 pKa = 11.52EE738 pKa = 4.02GGASIATFGLTGFISIVLLMLVV760 pKa = 3.79
MM1 pKa = 7.57KK2 pKa = 10.38LLSTVATAALLLQKK16 pKa = 10.49ARR18 pKa = 11.84SLTITEE24 pKa = 4.23DD25 pKa = 3.51TVLVSPVNLEE35 pKa = 3.67IGEE38 pKa = 4.44LNINPGVYY46 pKa = 10.31FSIVNNVLTVLGGNLNNDD64 pKa = 3.23GAFYY68 pKa = 9.77VTSTNGLAASVTIASGSIINRR89 pKa = 11.84GDD91 pKa = 3.29LAFNSLKK98 pKa = 11.12ANVITNFNLDD108 pKa = 3.45SVGTFSNTGNMWLGVPIFSAVPPIILGSALDD139 pKa = 3.87WEE141 pKa = 4.77NKK143 pKa = 8.73GMIYY147 pKa = 10.4LRR149 pKa = 11.84QEE151 pKa = 3.94LGGASPITISQVLGAIDD168 pKa = 3.82NSGTICIEE176 pKa = 3.84RR177 pKa = 11.84LNWLQTTTINGAGCVNVQADD197 pKa = 3.72GHH199 pKa = 6.3LQLQISPWSVGEE211 pKa = 4.0DD212 pKa = 3.09QTIYY216 pKa = 11.12LSTPTSALSVLGLEE230 pKa = 4.48PSLLGTKK237 pKa = 9.05TYY239 pKa = 11.13NVVGFGGGNTIGINLGFTSYY259 pKa = 10.56SYY261 pKa = 11.12SGSTLTLSFFLGVFKK276 pKa = 10.9INFNIGEE283 pKa = 4.58GYY285 pKa = 10.06SADD288 pKa = 3.37GFSTNGPGNSGTQITYY304 pKa = 10.21DD305 pKa = 3.37GPYY308 pKa = 9.31PGSVPDD314 pKa = 3.87KK315 pKa = 10.07CLCKK319 pKa = 10.57DD320 pKa = 3.9FPTPPTEE327 pKa = 4.35PSSSSSSSSSAPPSSSAPEE346 pKa = 3.86SSSTSEE352 pKa = 4.09SSSSEE357 pKa = 3.92SSSSSEE363 pKa = 4.02PPSSEE368 pKa = 4.07TSSSEE373 pKa = 4.15PPSSSQPPSSSEE385 pKa = 3.94PPSSSEE391 pKa = 4.42PPSSSQPPSSSQPPSSSEE409 pKa = 3.94PPSSSEE415 pKa = 4.42PPSSSQPPSSSEE427 pKa = 4.05PPSSSGPSEE436 pKa = 4.19SSGSSSGPSEE446 pKa = 4.2TSGSSSGPSEE456 pKa = 4.17TSGSSTGPSEE466 pKa = 3.93TSGPSEE472 pKa = 4.36SSTGPGNGGEE482 pKa = 4.23TSGPGNGGEE491 pKa = 4.28TSTGPGNGGQPTSGPGNGGEE511 pKa = 4.08PTGPGNGGEE520 pKa = 4.11PTGPGNGGEE529 pKa = 4.11PTGPGNGGQPTGPGNGGEE547 pKa = 4.11PTGPGNGGQPTGPGNGGQPTGPGNGGQPTGPGNGGQPTNGPGNGGQPTGPGGPSEE602 pKa = 4.09TAPGGGNGGGNGNGGGNGNGGGNGNGGGNGNGGEE636 pKa = 4.08NGNGGGNGNGGGNGNGGEE654 pKa = 4.07NGNGGEE660 pKa = 4.17NGNGGGNGTGGEE672 pKa = 4.07NGNGGGNGNGGGNGSGNGSGNGSGNGSGNGSGNGSGNGNGSGNGIEE718 pKa = 4.58TKK720 pKa = 10.03TSQSPTGGAGGPTVSSYY737 pKa = 11.52EE738 pKa = 4.02GGASIATFGLTGFISIVLLMLVV760 pKa = 3.79
Molecular weight: 72.27 kDa
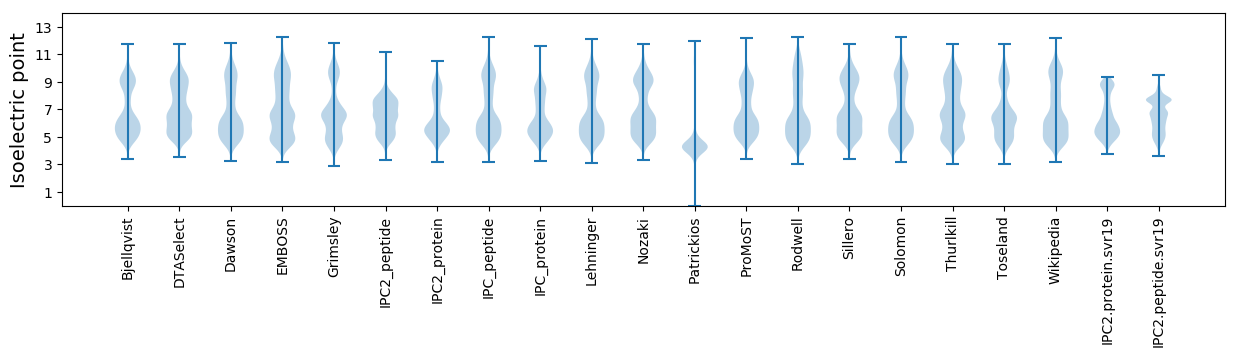
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H0ZCE9|A0A2H0ZCE9_CANAR Uncharacterized protein OS=Candida auris OX=498019 GN=B9J08_005011 PE=3 SV=1
MM1 pKa = 7.66IYY3 pKa = 9.12STTSRR8 pKa = 11.84LPAKK12 pKa = 10.52SKK14 pKa = 11.02GPLSKK19 pKa = 9.91RR20 pKa = 11.84TRR22 pKa = 11.84CRR24 pKa = 11.84HH25 pKa = 4.95FPYY28 pKa = 10.45SEE30 pKa = 4.36LKK32 pKa = 10.54LLHH35 pKa = 6.51TSTEE39 pKa = 4.03ATDD42 pKa = 3.07KK43 pKa = 10.73SKK45 pKa = 10.85RR46 pKa = 11.84YY47 pKa = 9.41RR48 pKa = 11.84PRR50 pKa = 11.84NQFMMARR57 pKa = 11.84QHH59 pKa = 5.63LTSTFKK65 pKa = 10.81DD66 pKa = 3.94GRR68 pKa = 11.84CARR71 pKa = 11.84VKK73 pKa = 10.38SIRR76 pKa = 11.84ASQVCSLSCRR86 pKa = 11.84FSSGLLRR93 pKa = 11.84NTVRR97 pKa = 11.84TKK99 pKa = 10.25FNCTVSMQYY108 pKa = 11.14SSIASCEE115 pKa = 4.0LSLQGCQISGAVEE128 pKa = 3.73TAGSKK133 pKa = 9.4KK134 pKa = 10.31RR135 pKa = 11.84KK136 pKa = 8.74IFLWLTGGRR145 pKa = 11.84CGTFSSMQILRR156 pKa = 11.84KK157 pKa = 9.92GG158 pKa = 3.23
MM1 pKa = 7.66IYY3 pKa = 9.12STTSRR8 pKa = 11.84LPAKK12 pKa = 10.52SKK14 pKa = 11.02GPLSKK19 pKa = 9.91RR20 pKa = 11.84TRR22 pKa = 11.84CRR24 pKa = 11.84HH25 pKa = 4.95FPYY28 pKa = 10.45SEE30 pKa = 4.36LKK32 pKa = 10.54LLHH35 pKa = 6.51TSTEE39 pKa = 4.03ATDD42 pKa = 3.07KK43 pKa = 10.73SKK45 pKa = 10.85RR46 pKa = 11.84YY47 pKa = 9.41RR48 pKa = 11.84PRR50 pKa = 11.84NQFMMARR57 pKa = 11.84QHH59 pKa = 5.63LTSTFKK65 pKa = 10.81DD66 pKa = 3.94GRR68 pKa = 11.84CARR71 pKa = 11.84VKK73 pKa = 10.38SIRR76 pKa = 11.84ASQVCSLSCRR86 pKa = 11.84FSSGLLRR93 pKa = 11.84NTVRR97 pKa = 11.84TKK99 pKa = 10.25FNCTVSMQYY108 pKa = 11.14SSIASCEE115 pKa = 4.0LSLQGCQISGAVEE128 pKa = 3.73TAGSKK133 pKa = 9.4KK134 pKa = 10.31RR135 pKa = 11.84KK136 pKa = 8.74IFLWLTGGRR145 pKa = 11.84CGTFSSMQILRR156 pKa = 11.84KK157 pKa = 9.92GG158 pKa = 3.23
Molecular weight: 17.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2694904 |
66 |
4946 |
498.2 |
55.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.699 ± 0.033 | 1.142 ± 0.012 |
5.899 ± 0.023 | 6.837 ± 0.037 |
4.398 ± 0.02 | 5.489 ± 0.033 |
2.327 ± 0.015 | 5.602 ± 0.021 |
6.878 ± 0.033 | 9.265 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.05 ± 0.01 | 4.839 ± 0.02 |
4.721 ± 0.029 | 3.932 ± 0.025 |
4.763 ± 0.023 | 9.177 ± 0.042 |
5.445 ± 0.041 | 6.217 ± 0.024 |
1.07 ± 0.01 | 3.249 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
