
Lactococcus phage phismq86
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
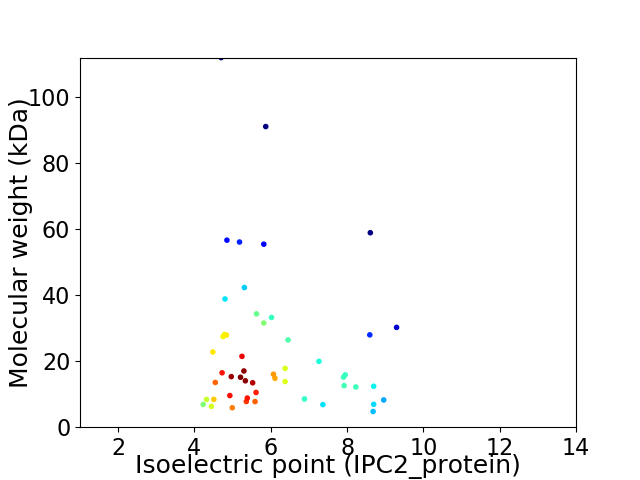
Virtual 2D-PAGE plot for 51 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A5GZ65|A5GZ65_9CAUD Putative repressor OS=Lactococcus phage phismq86 OX=444474 PE=4 SV=1
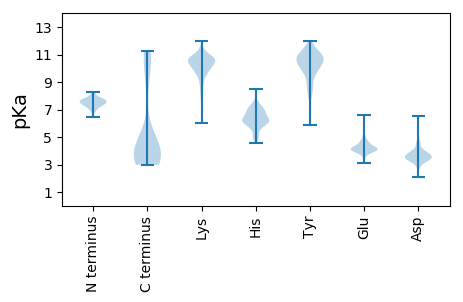
MM1 pKa = 8.24DD2 pKa = 4.03EE3 pKa = 4.4QEE5 pKa = 4.71LNSLLICEE13 pKa = 5.17IEE15 pKa = 4.2NQHH18 pKa = 6.03IDD20 pKa = 3.58YY21 pKa = 11.09RR22 pKa = 11.84LGNWNNQVAWVAPLLGLGGYY42 pKa = 7.42EE43 pKa = 3.92KK44 pKa = 10.65NARR47 pKa = 11.84PFDD50 pKa = 3.93HH51 pKa = 6.96AHH53 pKa = 6.01EE54 pKa = 4.77LSHH57 pKa = 7.34ILNHH61 pKa = 6.86DD62 pKa = 3.92DD63 pKa = 4.86YY64 pKa = 11.69RR65 pKa = 11.84GGDD68 pKa = 4.12CDD70 pKa = 3.54TTSPNEE76 pKa = 4.03SRR78 pKa = 11.84AHH80 pKa = 6.33RR81 pKa = 11.84EE82 pKa = 4.04AILLLWDD89 pKa = 3.55MFEE92 pKa = 4.48KK93 pKa = 10.72QGGDD97 pKa = 3.23YY98 pKa = 11.22SHH100 pKa = 7.41FNLFIEE106 pKa = 4.53ITGCPYY112 pKa = 11.09DD113 pKa = 3.48FAYY116 pKa = 10.86SIISKK121 pKa = 9.43EE122 pKa = 3.88FNEE125 pKa = 4.38MYY127 pKa = 10.42EE128 pKa = 4.43AINEE132 pKa = 4.15IFVDD136 pKa = 3.74EE137 pKa = 4.61LNIKK141 pKa = 9.16IKK143 pKa = 10.61KK144 pKa = 7.55EE145 pKa = 3.99QIHH148 pKa = 6.3KK149 pKa = 10.25FAVDD153 pKa = 3.52YY154 pKa = 10.83ISYY157 pKa = 10.64FDD159 pKa = 4.47IIEE162 pKa = 4.66SINIYY167 pKa = 10.81NFLEE171 pKa = 4.63AYY173 pKa = 9.72NLNHH177 pKa = 6.37SFYY180 pKa = 11.21DD181 pKa = 3.44LAEE184 pKa = 4.44RR185 pKa = 11.84EE186 pKa = 4.21FQEE189 pKa = 4.39LLGVAA194 pKa = 4.42
MM1 pKa = 8.24DD2 pKa = 4.03EE3 pKa = 4.4QEE5 pKa = 4.71LNSLLICEE13 pKa = 5.17IEE15 pKa = 4.2NQHH18 pKa = 6.03IDD20 pKa = 3.58YY21 pKa = 11.09RR22 pKa = 11.84LGNWNNQVAWVAPLLGLGGYY42 pKa = 7.42EE43 pKa = 3.92KK44 pKa = 10.65NARR47 pKa = 11.84PFDD50 pKa = 3.93HH51 pKa = 6.96AHH53 pKa = 6.01EE54 pKa = 4.77LSHH57 pKa = 7.34ILNHH61 pKa = 6.86DD62 pKa = 3.92DD63 pKa = 4.86YY64 pKa = 11.69RR65 pKa = 11.84GGDD68 pKa = 4.12CDD70 pKa = 3.54TTSPNEE76 pKa = 4.03SRR78 pKa = 11.84AHH80 pKa = 6.33RR81 pKa = 11.84EE82 pKa = 4.04AILLLWDD89 pKa = 3.55MFEE92 pKa = 4.48KK93 pKa = 10.72QGGDD97 pKa = 3.23YY98 pKa = 11.22SHH100 pKa = 7.41FNLFIEE106 pKa = 4.53ITGCPYY112 pKa = 11.09DD113 pKa = 3.48FAYY116 pKa = 10.86SIISKK121 pKa = 9.43EE122 pKa = 3.88FNEE125 pKa = 4.38MYY127 pKa = 10.42EE128 pKa = 4.43AINEE132 pKa = 4.15IFVDD136 pKa = 3.74EE137 pKa = 4.61LNIKK141 pKa = 9.16IKK143 pKa = 10.61KK144 pKa = 7.55EE145 pKa = 3.99QIHH148 pKa = 6.3KK149 pKa = 10.25FAVDD153 pKa = 3.52YY154 pKa = 10.83ISYY157 pKa = 10.64FDD159 pKa = 4.47IIEE162 pKa = 4.66SINIYY167 pKa = 10.81NFLEE171 pKa = 4.63AYY173 pKa = 9.72NLNHH177 pKa = 6.37SFYY180 pKa = 11.21DD181 pKa = 3.44LAEE184 pKa = 4.44RR185 pKa = 11.84EE186 pKa = 4.21FQEE189 pKa = 4.39LLGVAA194 pKa = 4.42
Molecular weight: 22.77 kDa
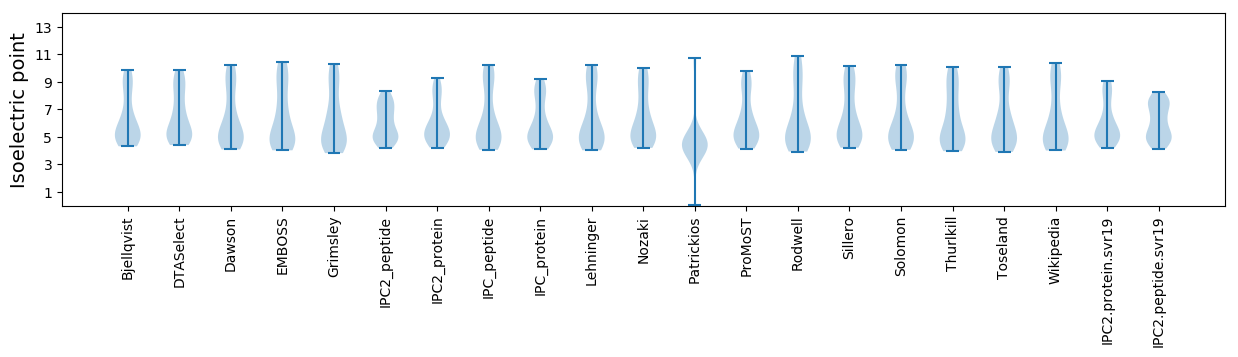
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A5GZ22|A5GZ22_9CAUD Uncharacterized protein OS=Lactococcus phage phismq86 OX=444474 PE=4 SV=1
MM1 pKa = 7.76RR2 pKa = 11.84GCFFVIIIYY11 pKa = 10.69SMNNIEE17 pKa = 4.97SVYY20 pKa = 10.77LNILRR25 pKa = 11.84GVKK28 pKa = 10.36NNLTNIIHH36 pKa = 6.54YY37 pKa = 8.36IQPNINIGDD46 pKa = 3.92VLSTTIALFALWYY59 pKa = 7.77TIHH62 pKa = 6.15STRR65 pKa = 11.84DD66 pKa = 3.43NQRR69 pKa = 11.84KK70 pKa = 8.93SVLPYY75 pKa = 7.97MTLEE79 pKa = 4.28QIRR82 pKa = 11.84STVTVDD88 pKa = 4.55GIGHH92 pKa = 6.04NTTVVDD98 pKa = 3.64TTSDD102 pKa = 3.48YY103 pKa = 11.11KK104 pKa = 11.47VRR106 pKa = 11.84VTKK109 pKa = 10.79NKK111 pKa = 8.09EE112 pKa = 3.81LKK114 pKa = 10.73GIEE117 pKa = 3.84VWGEE121 pKa = 3.85EE122 pKa = 3.99EE123 pKa = 4.09RR124 pKa = 11.84KK125 pKa = 10.48LIINEE130 pKa = 3.71KK131 pKa = 10.47SEE133 pKa = 4.35GKK135 pKa = 10.35GALSYY140 pKa = 11.43GSINFPRR147 pKa = 11.84SFKK150 pKa = 10.88LINTGKK156 pKa = 8.15NTAISFSLIVGKK168 pKa = 10.68KK169 pKa = 6.0STYY172 pKa = 9.57VKK174 pKa = 10.48NIAVNEE180 pKa = 4.04EE181 pKa = 4.24KK182 pKa = 10.33IISVLLEE189 pKa = 4.47DD190 pKa = 4.47EE191 pKa = 4.6NSTFNLLIKK200 pKa = 10.8FKK202 pKa = 10.52DD203 pKa = 3.28IYY205 pKa = 10.61GNQYY209 pKa = 9.39KK210 pKa = 10.32EE211 pKa = 4.03KK212 pKa = 9.84IEE214 pKa = 4.47FYY216 pKa = 9.67RR217 pKa = 11.84GRR219 pKa = 11.84MFLHH223 pKa = 7.12LDD225 pKa = 3.84LKK227 pKa = 10.76RR228 pKa = 11.84IRR230 pKa = 11.84FSKK233 pKa = 9.38TKK235 pKa = 10.36SIFMKK240 pKa = 9.45WKK242 pKa = 9.74SRR244 pKa = 11.84LKK246 pKa = 10.11IRR248 pKa = 11.84KK249 pKa = 6.66QARR252 pKa = 11.84NEE254 pKa = 3.8KK255 pKa = 10.55NKK257 pKa = 10.26LDD259 pKa = 3.72KK260 pKa = 11.23
MM1 pKa = 7.76RR2 pKa = 11.84GCFFVIIIYY11 pKa = 10.69SMNNIEE17 pKa = 4.97SVYY20 pKa = 10.77LNILRR25 pKa = 11.84GVKK28 pKa = 10.36NNLTNIIHH36 pKa = 6.54YY37 pKa = 8.36IQPNINIGDD46 pKa = 3.92VLSTTIALFALWYY59 pKa = 7.77TIHH62 pKa = 6.15STRR65 pKa = 11.84DD66 pKa = 3.43NQRR69 pKa = 11.84KK70 pKa = 8.93SVLPYY75 pKa = 7.97MTLEE79 pKa = 4.28QIRR82 pKa = 11.84STVTVDD88 pKa = 4.55GIGHH92 pKa = 6.04NTTVVDD98 pKa = 3.64TTSDD102 pKa = 3.48YY103 pKa = 11.11KK104 pKa = 11.47VRR106 pKa = 11.84VTKK109 pKa = 10.79NKK111 pKa = 8.09EE112 pKa = 3.81LKK114 pKa = 10.73GIEE117 pKa = 3.84VWGEE121 pKa = 3.85EE122 pKa = 3.99EE123 pKa = 4.09RR124 pKa = 11.84KK125 pKa = 10.48LIINEE130 pKa = 3.71KK131 pKa = 10.47SEE133 pKa = 4.35GKK135 pKa = 10.35GALSYY140 pKa = 11.43GSINFPRR147 pKa = 11.84SFKK150 pKa = 10.88LINTGKK156 pKa = 8.15NTAISFSLIVGKK168 pKa = 10.68KK169 pKa = 6.0STYY172 pKa = 9.57VKK174 pKa = 10.48NIAVNEE180 pKa = 4.04EE181 pKa = 4.24KK182 pKa = 10.33IISVLLEE189 pKa = 4.47DD190 pKa = 4.47EE191 pKa = 4.6NSTFNLLIKK200 pKa = 10.8FKK202 pKa = 10.52DD203 pKa = 3.28IYY205 pKa = 10.61GNQYY209 pKa = 9.39KK210 pKa = 10.32EE211 pKa = 4.03KK212 pKa = 9.84IEE214 pKa = 4.47FYY216 pKa = 9.67RR217 pKa = 11.84GRR219 pKa = 11.84MFLHH223 pKa = 7.12LDD225 pKa = 3.84LKK227 pKa = 10.76RR228 pKa = 11.84IRR230 pKa = 11.84FSKK233 pKa = 9.38TKK235 pKa = 10.36SIFMKK240 pKa = 9.45WKK242 pKa = 9.74SRR244 pKa = 11.84LKK246 pKa = 10.11IRR248 pKa = 11.84KK249 pKa = 6.66QARR252 pKa = 11.84NEE254 pKa = 3.8KK255 pKa = 10.55NKK257 pKa = 10.26LDD259 pKa = 3.72KK260 pKa = 11.23
Molecular weight: 30.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10604 |
40 |
1027 |
207.9 |
23.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.469 ± 0.455 | 0.538 ± 0.153 |
6.375 ± 0.254 | 7.422 ± 0.793 |
4.14 ± 0.219 | 6.582 ± 0.463 |
1.358 ± 0.165 | 7.318 ± 0.499 |
8.223 ± 0.484 | 7.912 ± 0.419 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.48 ± 0.274 | 5.828 ± 0.281 |
2.942 ± 0.187 | 4.121 ± 0.221 |
3.838 ± 0.356 | 6.846 ± 0.339 |
6.884 ± 0.69 | 5.988 ± 0.242 |
1.245 ± 0.138 | 3.489 ± 0.238 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
