
Blotched snakehead virus (BSNV) (Channa lucius virus)
Taxonomy: Viruses; Riboviria; Orthornavirae; Birnaviridae; Blosnavirus
Average proteome isoelectric point is 7.18
Get precalculated fractions of proteins
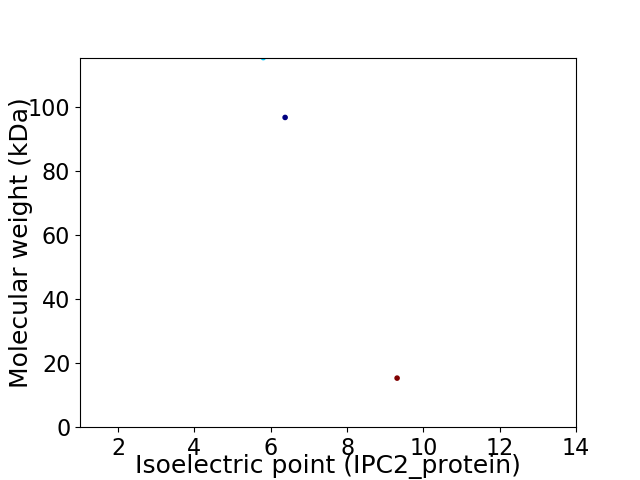
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q8AZM0|POLS_BSNV Structural polyprotein OS=Blotched snakehead virus OX=311176 PE=1 SV=1
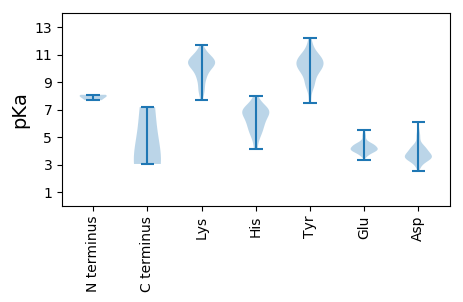
MM1 pKa = 7.97DD2 pKa = 4.58FSKK5 pKa = 11.13EE6 pKa = 3.92NTQIRR11 pKa = 11.84YY12 pKa = 9.54LNSLLVPEE20 pKa = 5.0TGSTSIPDD28 pKa = 3.52DD29 pKa = 3.54TLDD32 pKa = 3.49RR33 pKa = 11.84HH34 pKa = 5.58CLKK37 pKa = 10.25TEE39 pKa = 3.93TTTEE43 pKa = 3.98NLVAALGGSGLIVLFPNSPSGLLGAHH69 pKa = 5.23YY70 pKa = 8.57TKK72 pKa = 10.3TPQGSLIFDD81 pKa = 3.8KK82 pKa = 11.25AITTSQDD89 pKa = 2.56LKK91 pKa = 10.71KK92 pKa = 10.38AYY94 pKa = 10.34NYY96 pKa = 10.35ARR98 pKa = 11.84LVSRR102 pKa = 11.84IVQVRR107 pKa = 11.84SSTLPAGVYY116 pKa = 10.3ALNGTFNGVTYY127 pKa = 10.33IGSLSEE133 pKa = 5.5IKK135 pKa = 10.78DD136 pKa = 3.53LDD138 pKa = 4.14YY139 pKa = 11.78NSLLSATANINDD151 pKa = 3.56KK152 pKa = 10.83VGNVLVGDD160 pKa = 4.49GVAVLSLPAGSDD172 pKa = 3.37LPYY175 pKa = 10.7VRR177 pKa = 11.84LGDD180 pKa = 4.15EE181 pKa = 4.63VPSSAGVARR190 pKa = 11.84CSPSDD195 pKa = 3.44RR196 pKa = 11.84PRR198 pKa = 11.84HH199 pKa = 4.92YY200 pKa = 10.67NANNKK205 pKa = 7.72QVQVGTTDD213 pKa = 2.94TKK215 pKa = 11.11TNGFNIDD222 pKa = 3.12ATTPTEE228 pKa = 4.12VTVDD232 pKa = 3.41MQIAQIAAGKK242 pKa = 7.69TLTVTVKK249 pKa = 10.8LMGLTGAKK257 pKa = 9.11VASRR261 pKa = 11.84SEE263 pKa = 4.38TVSGNGGTFHH273 pKa = 7.61FSTTAVFGEE282 pKa = 4.5TEE284 pKa = 3.6ITQPVVGVQVLAKK297 pKa = 9.84TNGDD301 pKa = 4.14PIVVDD306 pKa = 3.51SYY308 pKa = 11.87VGVTVHH314 pKa = 6.55GGNMPGTLRR323 pKa = 11.84PVTIIAYY330 pKa = 9.09EE331 pKa = 4.26SVATGSVLTLSGISNYY347 pKa = 8.83EE348 pKa = 4.17LIPNPEE354 pKa = 4.06LAKK357 pKa = 10.68NIQTSYY363 pKa = 11.25GKK365 pKa = 10.58LNPAEE370 pKa = 3.92MTYY373 pKa = 10.14TKK375 pKa = 10.69VVLSHH380 pKa = 7.01RR381 pKa = 11.84DD382 pKa = 3.52EE383 pKa = 5.0LGLRR387 pKa = 11.84SIWSIPQYY395 pKa = 10.64RR396 pKa = 11.84DD397 pKa = 2.79MMSYY401 pKa = 10.06FRR403 pKa = 11.84EE404 pKa = 4.02VSDD407 pKa = 3.74RR408 pKa = 11.84SSPLKK413 pKa = 10.33IAGAFGWGDD422 pKa = 3.76LLSGIRR428 pKa = 11.84KK429 pKa = 8.19WVFPVVDD436 pKa = 3.95TLLPAARR443 pKa = 11.84PLTDD447 pKa = 5.16LASGWIKK454 pKa = 10.94NKK456 pKa = 10.24YY457 pKa = 8.75PEE459 pKa = 4.45AASGRR464 pKa = 11.84PLAASGRR471 pKa = 11.84PMAASGTFSKK481 pKa = 10.64RR482 pKa = 11.84IPLASSDD489 pKa = 3.95EE490 pKa = 3.88IDD492 pKa = 4.02YY493 pKa = 11.33QSVLALTIPGTHH505 pKa = 7.19PKK507 pKa = 9.98LVPPTEE513 pKa = 4.33RR514 pKa = 11.84EE515 pKa = 4.17PNSTPDD521 pKa = 2.95GHH523 pKa = 6.98KK524 pKa = 9.43ITGAKK529 pKa = 9.08TKK531 pKa = 10.59DD532 pKa = 3.32NTGGDD537 pKa = 3.71VTVVKK542 pKa = 10.36PLDD545 pKa = 3.49WLFKK549 pKa = 10.43LPCLRR554 pKa = 11.84PQAADD559 pKa = 3.71LPISLLQTLAYY570 pKa = 8.74KK571 pKa = 10.46QPLGRR576 pKa = 11.84NSRR579 pKa = 11.84IVHH582 pKa = 5.47FTDD585 pKa = 4.02GALFPVVAFGDD596 pKa = 3.53NHH598 pKa = 5.9STSEE602 pKa = 4.03LYY604 pKa = 9.93IAVRR608 pKa = 11.84GDD610 pKa = 3.33HH611 pKa = 7.29RR612 pKa = 11.84DD613 pKa = 3.32LMSPDD618 pKa = 3.34VRR620 pKa = 11.84DD621 pKa = 3.83SYY623 pKa = 12.17ALTGDD628 pKa = 3.69DD629 pKa = 3.76HH630 pKa = 6.89KK631 pKa = 11.7VWGATHH637 pKa = 5.52HH638 pKa = 6.08TYY640 pKa = 10.59YY641 pKa = 10.99VEE643 pKa = 4.44GAPKK647 pKa = 10.27KK648 pKa = 9.48PLKK651 pKa = 10.76FNVKK655 pKa = 8.37TRR657 pKa = 11.84TDD659 pKa = 3.54LTILPVADD667 pKa = 3.74VFWRR671 pKa = 11.84ADD673 pKa = 3.02GSADD677 pKa = 4.27VDD679 pKa = 4.38VVWNDD684 pKa = 3.21MPAVAGQSSSIALALASSLPFVPKK708 pKa = 10.33AAYY711 pKa = 7.66TGCLSGTNVQPVQFGNLKK729 pKa = 10.06ARR731 pKa = 11.84AAHH734 pKa = 6.96KK735 pKa = 10.35IGLPLVGMTQDD746 pKa = 3.14GGEE749 pKa = 4.19DD750 pKa = 3.48TRR752 pKa = 11.84ICTLDD757 pKa = 3.71DD758 pKa = 4.25AADD761 pKa = 3.59HH762 pKa = 6.81AFDD765 pKa = 4.33SMEE768 pKa = 4.18STVTRR773 pKa = 11.84PEE775 pKa = 4.34SVGHH779 pKa = 5.22QAAFQGWFYY788 pKa = 11.13CGAADD793 pKa = 4.36EE794 pKa = 4.42EE795 pKa = 4.87TIEE798 pKa = 4.11EE799 pKa = 4.7LEE801 pKa = 4.74DD802 pKa = 3.82FLDD805 pKa = 4.9SIEE808 pKa = 4.36LHH810 pKa = 6.33SKK812 pKa = 8.3PTVEE816 pKa = 4.44QPQTEE821 pKa = 4.24EE822 pKa = 3.9AMEE825 pKa = 4.3LLMEE829 pKa = 5.13LARR832 pKa = 11.84KK833 pKa = 9.16DD834 pKa = 3.55PQMSKK839 pKa = 10.25ILVILGWVEE848 pKa = 3.82GAGLIDD854 pKa = 5.41ALYY857 pKa = 10.93NWAQLDD863 pKa = 3.84DD864 pKa = 4.18GGVRR868 pKa = 11.84MRR870 pKa = 11.84NMLRR874 pKa = 11.84NLPHH878 pKa = 6.97EE879 pKa = 4.72GSKK882 pKa = 9.04SQRR885 pKa = 11.84RR886 pKa = 11.84KK887 pKa = 10.05HH888 pKa = 5.77GPAPEE893 pKa = 4.24SRR895 pKa = 11.84EE896 pKa = 4.06STRR899 pKa = 11.84MEE901 pKa = 3.99VLRR904 pKa = 11.84RR905 pKa = 11.84EE906 pKa = 4.15AAAKK910 pKa = 9.78RR911 pKa = 11.84KK912 pKa = 8.53KK913 pKa = 10.01AQRR916 pKa = 11.84ISEE919 pKa = 4.16DD920 pKa = 3.19AMDD923 pKa = 4.34NGFEE927 pKa = 4.16FATIDD932 pKa = 3.22WVLEE936 pKa = 4.04NGSRR940 pKa = 11.84GPNPAQAKK948 pKa = 8.52YY949 pKa = 10.62YY950 pKa = 10.15KK951 pKa = 9.92ATGLDD956 pKa = 3.86PEE958 pKa = 5.19PGLTEE963 pKa = 4.17FLPEE967 pKa = 3.89PTHH970 pKa = 7.63APEE973 pKa = 4.03NKK975 pKa = 9.32AAKK978 pKa = 9.59LAATIYY984 pKa = 10.23GSPNQAPAPPEE995 pKa = 3.79FVEE998 pKa = 4.67EE999 pKa = 4.15VAAVLMEE1006 pKa = 4.37NNGRR1010 pKa = 11.84GPNQAQMRR1018 pKa = 11.84EE1019 pKa = 4.17LRR1021 pKa = 11.84LKK1023 pKa = 10.66ALTMKK1028 pKa = 10.32SGSGAAATFKK1038 pKa = 10.5PRR1040 pKa = 11.84NRR1042 pKa = 11.84RR1043 pKa = 11.84PAQEE1047 pKa = 3.99YY1048 pKa = 9.21QPRR1051 pKa = 11.84PPITSRR1057 pKa = 11.84AGRR1060 pKa = 11.84FLNISTTLSS1069 pKa = 3.19
MM1 pKa = 7.97DD2 pKa = 4.58FSKK5 pKa = 11.13EE6 pKa = 3.92NTQIRR11 pKa = 11.84YY12 pKa = 9.54LNSLLVPEE20 pKa = 5.0TGSTSIPDD28 pKa = 3.52DD29 pKa = 3.54TLDD32 pKa = 3.49RR33 pKa = 11.84HH34 pKa = 5.58CLKK37 pKa = 10.25TEE39 pKa = 3.93TTTEE43 pKa = 3.98NLVAALGGSGLIVLFPNSPSGLLGAHH69 pKa = 5.23YY70 pKa = 8.57TKK72 pKa = 10.3TPQGSLIFDD81 pKa = 3.8KK82 pKa = 11.25AITTSQDD89 pKa = 2.56LKK91 pKa = 10.71KK92 pKa = 10.38AYY94 pKa = 10.34NYY96 pKa = 10.35ARR98 pKa = 11.84LVSRR102 pKa = 11.84IVQVRR107 pKa = 11.84SSTLPAGVYY116 pKa = 10.3ALNGTFNGVTYY127 pKa = 10.33IGSLSEE133 pKa = 5.5IKK135 pKa = 10.78DD136 pKa = 3.53LDD138 pKa = 4.14YY139 pKa = 11.78NSLLSATANINDD151 pKa = 3.56KK152 pKa = 10.83VGNVLVGDD160 pKa = 4.49GVAVLSLPAGSDD172 pKa = 3.37LPYY175 pKa = 10.7VRR177 pKa = 11.84LGDD180 pKa = 4.15EE181 pKa = 4.63VPSSAGVARR190 pKa = 11.84CSPSDD195 pKa = 3.44RR196 pKa = 11.84PRR198 pKa = 11.84HH199 pKa = 4.92YY200 pKa = 10.67NANNKK205 pKa = 7.72QVQVGTTDD213 pKa = 2.94TKK215 pKa = 11.11TNGFNIDD222 pKa = 3.12ATTPTEE228 pKa = 4.12VTVDD232 pKa = 3.41MQIAQIAAGKK242 pKa = 7.69TLTVTVKK249 pKa = 10.8LMGLTGAKK257 pKa = 9.11VASRR261 pKa = 11.84SEE263 pKa = 4.38TVSGNGGTFHH273 pKa = 7.61FSTTAVFGEE282 pKa = 4.5TEE284 pKa = 3.6ITQPVVGVQVLAKK297 pKa = 9.84TNGDD301 pKa = 4.14PIVVDD306 pKa = 3.51SYY308 pKa = 11.87VGVTVHH314 pKa = 6.55GGNMPGTLRR323 pKa = 11.84PVTIIAYY330 pKa = 9.09EE331 pKa = 4.26SVATGSVLTLSGISNYY347 pKa = 8.83EE348 pKa = 4.17LIPNPEE354 pKa = 4.06LAKK357 pKa = 10.68NIQTSYY363 pKa = 11.25GKK365 pKa = 10.58LNPAEE370 pKa = 3.92MTYY373 pKa = 10.14TKK375 pKa = 10.69VVLSHH380 pKa = 7.01RR381 pKa = 11.84DD382 pKa = 3.52EE383 pKa = 5.0LGLRR387 pKa = 11.84SIWSIPQYY395 pKa = 10.64RR396 pKa = 11.84DD397 pKa = 2.79MMSYY401 pKa = 10.06FRR403 pKa = 11.84EE404 pKa = 4.02VSDD407 pKa = 3.74RR408 pKa = 11.84SSPLKK413 pKa = 10.33IAGAFGWGDD422 pKa = 3.76LLSGIRR428 pKa = 11.84KK429 pKa = 8.19WVFPVVDD436 pKa = 3.95TLLPAARR443 pKa = 11.84PLTDD447 pKa = 5.16LASGWIKK454 pKa = 10.94NKK456 pKa = 10.24YY457 pKa = 8.75PEE459 pKa = 4.45AASGRR464 pKa = 11.84PLAASGRR471 pKa = 11.84PMAASGTFSKK481 pKa = 10.64RR482 pKa = 11.84IPLASSDD489 pKa = 3.95EE490 pKa = 3.88IDD492 pKa = 4.02YY493 pKa = 11.33QSVLALTIPGTHH505 pKa = 7.19PKK507 pKa = 9.98LVPPTEE513 pKa = 4.33RR514 pKa = 11.84EE515 pKa = 4.17PNSTPDD521 pKa = 2.95GHH523 pKa = 6.98KK524 pKa = 9.43ITGAKK529 pKa = 9.08TKK531 pKa = 10.59DD532 pKa = 3.32NTGGDD537 pKa = 3.71VTVVKK542 pKa = 10.36PLDD545 pKa = 3.49WLFKK549 pKa = 10.43LPCLRR554 pKa = 11.84PQAADD559 pKa = 3.71LPISLLQTLAYY570 pKa = 8.74KK571 pKa = 10.46QPLGRR576 pKa = 11.84NSRR579 pKa = 11.84IVHH582 pKa = 5.47FTDD585 pKa = 4.02GALFPVVAFGDD596 pKa = 3.53NHH598 pKa = 5.9STSEE602 pKa = 4.03LYY604 pKa = 9.93IAVRR608 pKa = 11.84GDD610 pKa = 3.33HH611 pKa = 7.29RR612 pKa = 11.84DD613 pKa = 3.32LMSPDD618 pKa = 3.34VRR620 pKa = 11.84DD621 pKa = 3.83SYY623 pKa = 12.17ALTGDD628 pKa = 3.69DD629 pKa = 3.76HH630 pKa = 6.89KK631 pKa = 11.7VWGATHH637 pKa = 5.52HH638 pKa = 6.08TYY640 pKa = 10.59YY641 pKa = 10.99VEE643 pKa = 4.44GAPKK647 pKa = 10.27KK648 pKa = 9.48PLKK651 pKa = 10.76FNVKK655 pKa = 8.37TRR657 pKa = 11.84TDD659 pKa = 3.54LTILPVADD667 pKa = 3.74VFWRR671 pKa = 11.84ADD673 pKa = 3.02GSADD677 pKa = 4.27VDD679 pKa = 4.38VVWNDD684 pKa = 3.21MPAVAGQSSSIALALASSLPFVPKK708 pKa = 10.33AAYY711 pKa = 7.66TGCLSGTNVQPVQFGNLKK729 pKa = 10.06ARR731 pKa = 11.84AAHH734 pKa = 6.96KK735 pKa = 10.35IGLPLVGMTQDD746 pKa = 3.14GGEE749 pKa = 4.19DD750 pKa = 3.48TRR752 pKa = 11.84ICTLDD757 pKa = 3.71DD758 pKa = 4.25AADD761 pKa = 3.59HH762 pKa = 6.81AFDD765 pKa = 4.33SMEE768 pKa = 4.18STVTRR773 pKa = 11.84PEE775 pKa = 4.34SVGHH779 pKa = 5.22QAAFQGWFYY788 pKa = 11.13CGAADD793 pKa = 4.36EE794 pKa = 4.42EE795 pKa = 4.87TIEE798 pKa = 4.11EE799 pKa = 4.7LEE801 pKa = 4.74DD802 pKa = 3.82FLDD805 pKa = 4.9SIEE808 pKa = 4.36LHH810 pKa = 6.33SKK812 pKa = 8.3PTVEE816 pKa = 4.44QPQTEE821 pKa = 4.24EE822 pKa = 3.9AMEE825 pKa = 4.3LLMEE829 pKa = 5.13LARR832 pKa = 11.84KK833 pKa = 9.16DD834 pKa = 3.55PQMSKK839 pKa = 10.25ILVILGWVEE848 pKa = 3.82GAGLIDD854 pKa = 5.41ALYY857 pKa = 10.93NWAQLDD863 pKa = 3.84DD864 pKa = 4.18GGVRR868 pKa = 11.84MRR870 pKa = 11.84NMLRR874 pKa = 11.84NLPHH878 pKa = 6.97EE879 pKa = 4.72GSKK882 pKa = 9.04SQRR885 pKa = 11.84RR886 pKa = 11.84KK887 pKa = 10.05HH888 pKa = 5.77GPAPEE893 pKa = 4.24SRR895 pKa = 11.84EE896 pKa = 4.06STRR899 pKa = 11.84MEE901 pKa = 3.99VLRR904 pKa = 11.84RR905 pKa = 11.84EE906 pKa = 4.15AAAKK910 pKa = 9.78RR911 pKa = 11.84KK912 pKa = 8.53KK913 pKa = 10.01AQRR916 pKa = 11.84ISEE919 pKa = 4.16DD920 pKa = 3.19AMDD923 pKa = 4.34NGFEE927 pKa = 4.16FATIDD932 pKa = 3.22WVLEE936 pKa = 4.04NGSRR940 pKa = 11.84GPNPAQAKK948 pKa = 8.52YY949 pKa = 10.62YY950 pKa = 10.15KK951 pKa = 9.92ATGLDD956 pKa = 3.86PEE958 pKa = 5.19PGLTEE963 pKa = 4.17FLPEE967 pKa = 3.89PTHH970 pKa = 7.63APEE973 pKa = 4.03NKK975 pKa = 9.32AAKK978 pKa = 9.59LAATIYY984 pKa = 10.23GSPNQAPAPPEE995 pKa = 3.79FVEE998 pKa = 4.67EE999 pKa = 4.15VAAVLMEE1006 pKa = 4.37NNGRR1010 pKa = 11.84GPNQAQMRR1018 pKa = 11.84EE1019 pKa = 4.17LRR1021 pKa = 11.84LKK1023 pKa = 10.66ALTMKK1028 pKa = 10.32SGSGAAATFKK1038 pKa = 10.5PRR1040 pKa = 11.84NRR1042 pKa = 11.84RR1043 pKa = 11.84PAQEE1047 pKa = 3.99YY1048 pKa = 9.21QPRR1051 pKa = 11.84PPITSRR1057 pKa = 11.84AGRR1060 pKa = 11.84FLNISTTLSS1069 pKa = 3.19
Molecular weight: 115.49 kDa
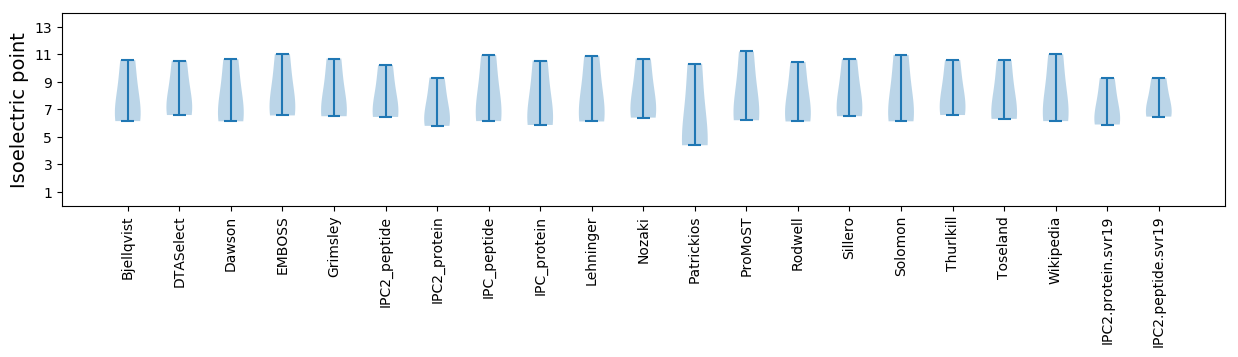
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q8AZM0|POLS_BSNV Structural polyprotein OS=Blotched snakehead virus OX=311176 PE=1 SV=1
MM1 pKa = 8.07PDD3 pKa = 3.86LSAALCRR10 pKa = 11.84CGAQRR15 pKa = 11.84SPRR18 pKa = 11.84VSTHH22 pKa = 4.16STGPSMGLPTLEE34 pKa = 3.64VSARR38 pKa = 11.84SRR40 pKa = 11.84TWTTIHH46 pKa = 7.04FSLQRR51 pKa = 11.84PTSMTRR57 pKa = 11.84SAMYY61 pKa = 10.69LLEE64 pKa = 4.3TVSQCSHH71 pKa = 6.83FLLEE75 pKa = 4.55ATCLMLDD82 pKa = 4.34LAMKK86 pKa = 9.51FQALLEE92 pKa = 4.47SRR94 pKa = 11.84DD95 pKa = 3.93ALPPTDD101 pKa = 5.49LGTTMPTTNRR111 pKa = 11.84SRR113 pKa = 11.84SARR116 pKa = 11.84PTPRR120 pKa = 11.84PMGSTSTQQHH130 pKa = 5.35PQRR133 pKa = 11.84SRR135 pKa = 11.84STCRR139 pKa = 3.04
MM1 pKa = 8.07PDD3 pKa = 3.86LSAALCRR10 pKa = 11.84CGAQRR15 pKa = 11.84SPRR18 pKa = 11.84VSTHH22 pKa = 4.16STGPSMGLPTLEE34 pKa = 3.64VSARR38 pKa = 11.84SRR40 pKa = 11.84TWTTIHH46 pKa = 7.04FSLQRR51 pKa = 11.84PTSMTRR57 pKa = 11.84SAMYY61 pKa = 10.69LLEE64 pKa = 4.3TVSQCSHH71 pKa = 6.83FLLEE75 pKa = 4.55ATCLMLDD82 pKa = 4.34LAMKK86 pKa = 9.51FQALLEE92 pKa = 4.47SRR94 pKa = 11.84DD95 pKa = 3.93ALPPTDD101 pKa = 5.49LGTTMPTTNRR111 pKa = 11.84SRR113 pKa = 11.84SARR116 pKa = 11.84PTPRR120 pKa = 11.84PMGSTSTQQHH130 pKa = 5.35PQRR133 pKa = 11.84SRR135 pKa = 11.84STCRR139 pKa = 3.04
Molecular weight: 15.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2075 |
139 |
1069 |
691.7 |
75.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.578 ± 0.598 | 0.867 ± 0.631 |
5.205 ± 0.604 | 5.735 ± 0.837 |
2.795 ± 0.148 | 7.181 ± 0.915 |
1.976 ± 0.21 | 3.904 ± 0.733 |
5.301 ± 1.164 | 9.976 ± 0.531 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.747 ± 0.758 | 4.53 ± 0.957 |
6.699 ± 0.477 | 3.422 ± 0.38 |
5.446 ± 1.228 | 7.47 ± 1.454 |
8.241 ± 1.246 | 5.735 ± 1.144 |
1.446 ± 0.297 | 2.747 ± 0.469 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
