
Bovine faeces associated smacovirus 4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Cosmacovirus; Cosmacovirus bovas1
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
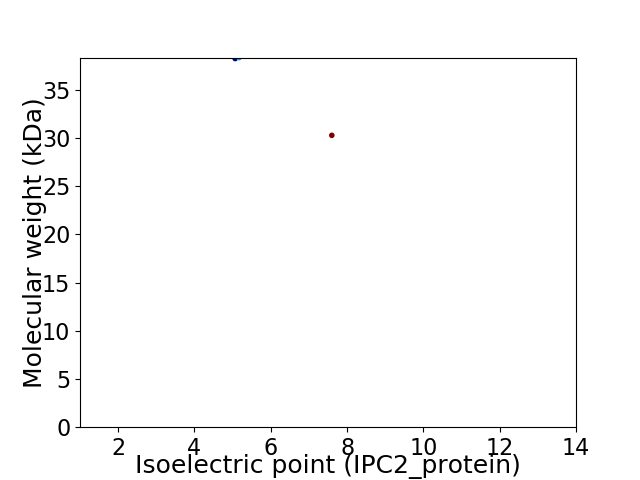
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
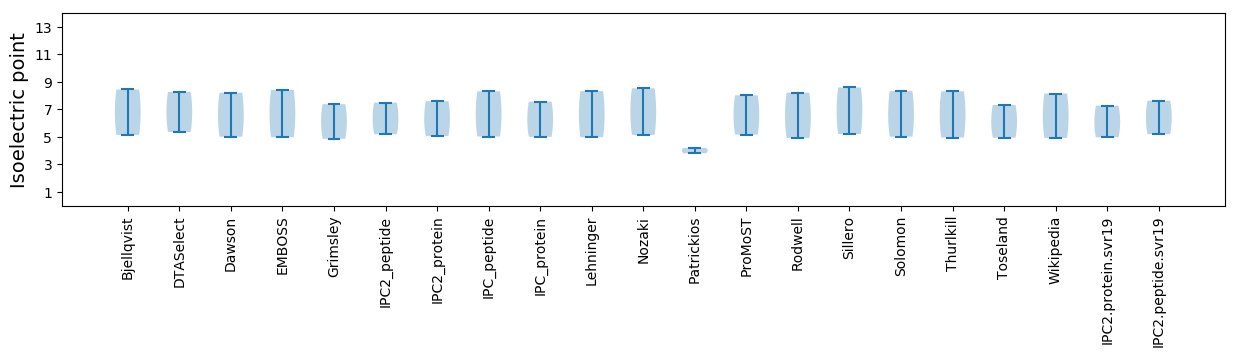
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A160HX24|A0A160HX24_9VIRU Replication associated protein OS=Bovine faeces associated smacovirus 4 OX=1843752 PE=4 SV=1
MM1 pKa = 6.36VTVTVSEE8 pKa = 4.52TYY10 pKa = 10.75DD11 pKa = 3.64LSTKK15 pKa = 8.29VNKK18 pKa = 9.5MSLIGIHH25 pKa = 5.95TPGKK29 pKa = 10.29SLIQKK34 pKa = 9.0SYY36 pKa = 10.79PGLLMNSKK44 pKa = 10.25YY45 pKa = 10.72ISIDD49 pKa = 3.36KK50 pKa = 10.88VDD52 pKa = 3.5VTLAAVSTLPISPDD66 pKa = 3.41QVGLDD71 pKa = 3.66VDD73 pKa = 4.88NIAPQDD79 pKa = 3.59MMNPILYY86 pKa = 9.82KK87 pKa = 10.58AVSNDD92 pKa = 2.82SMSNIEE98 pKa = 4.29ARR100 pKa = 11.84LHH102 pKa = 5.49GLGFGSGTQVSGSMVDD118 pKa = 4.19AINDD122 pKa = 3.59NVTGISDD129 pKa = 3.95EE130 pKa = 4.36FNVYY134 pKa = 10.57YY135 pKa = 10.74SLLSNRR141 pKa = 11.84DD142 pKa = 3.17GFRR145 pKa = 11.84IAHH148 pKa = 5.69PQQGLSMRR156 pKa = 11.84GLVPLVFDD164 pKa = 4.16KK165 pKa = 11.06YY166 pKa = 9.17YY167 pKa = 11.09NVGAFMTNSQEE178 pKa = 4.11VTDD181 pKa = 4.24VGDD184 pKa = 3.59TGADD188 pKa = 3.48EE189 pKa = 4.52TPSGDD194 pKa = 4.48LYY196 pKa = 10.31PTLQYY201 pKa = 11.08SAGSLSVVNRR211 pKa = 11.84PVQAMRR217 pKa = 11.84GRR219 pKa = 11.84PHH221 pKa = 7.69RR222 pKa = 11.84MPRR225 pKa = 11.84FPTLYY230 pKa = 8.86VTAASSGSSQSTNQRR245 pKa = 11.84VANGIADD252 pKa = 4.21GQPLNCEE259 pKa = 3.92VLMPDD264 pKa = 4.01ILPVYY269 pKa = 9.86LGMIIMPPAKK279 pKa = 8.57RR280 pKa = 11.84TVMFYY285 pKa = 11.05RR286 pKa = 11.84MVVRR290 pKa = 11.84ATLTFSEE297 pKa = 4.57VRR299 pKa = 11.84PMSEE303 pKa = 3.39ITSFAGLNSGYY314 pKa = 10.5GSEE317 pKa = 4.98VYY319 pKa = 8.8HH320 pKa = 6.71TDD322 pKa = 2.93YY323 pKa = 11.66ALQSKK328 pKa = 9.01QMEE331 pKa = 4.67STTDD335 pKa = 3.58LVDD338 pKa = 3.45VQNANIEE345 pKa = 4.31KK346 pKa = 10.25IMEE349 pKa = 4.16GRR351 pKa = 3.45
MM1 pKa = 6.36VTVTVSEE8 pKa = 4.52TYY10 pKa = 10.75DD11 pKa = 3.64LSTKK15 pKa = 8.29VNKK18 pKa = 9.5MSLIGIHH25 pKa = 5.95TPGKK29 pKa = 10.29SLIQKK34 pKa = 9.0SYY36 pKa = 10.79PGLLMNSKK44 pKa = 10.25YY45 pKa = 10.72ISIDD49 pKa = 3.36KK50 pKa = 10.88VDD52 pKa = 3.5VTLAAVSTLPISPDD66 pKa = 3.41QVGLDD71 pKa = 3.66VDD73 pKa = 4.88NIAPQDD79 pKa = 3.59MMNPILYY86 pKa = 9.82KK87 pKa = 10.58AVSNDD92 pKa = 2.82SMSNIEE98 pKa = 4.29ARR100 pKa = 11.84LHH102 pKa = 5.49GLGFGSGTQVSGSMVDD118 pKa = 4.19AINDD122 pKa = 3.59NVTGISDD129 pKa = 3.95EE130 pKa = 4.36FNVYY134 pKa = 10.57YY135 pKa = 10.74SLLSNRR141 pKa = 11.84DD142 pKa = 3.17GFRR145 pKa = 11.84IAHH148 pKa = 5.69PQQGLSMRR156 pKa = 11.84GLVPLVFDD164 pKa = 4.16KK165 pKa = 11.06YY166 pKa = 9.17YY167 pKa = 11.09NVGAFMTNSQEE178 pKa = 4.11VTDD181 pKa = 4.24VGDD184 pKa = 3.59TGADD188 pKa = 3.48EE189 pKa = 4.52TPSGDD194 pKa = 4.48LYY196 pKa = 10.31PTLQYY201 pKa = 11.08SAGSLSVVNRR211 pKa = 11.84PVQAMRR217 pKa = 11.84GRR219 pKa = 11.84PHH221 pKa = 7.69RR222 pKa = 11.84MPRR225 pKa = 11.84FPTLYY230 pKa = 8.86VTAASSGSSQSTNQRR245 pKa = 11.84VANGIADD252 pKa = 4.21GQPLNCEE259 pKa = 3.92VLMPDD264 pKa = 4.01ILPVYY269 pKa = 9.86LGMIIMPPAKK279 pKa = 8.57RR280 pKa = 11.84TVMFYY285 pKa = 11.05RR286 pKa = 11.84MVVRR290 pKa = 11.84ATLTFSEE297 pKa = 4.57VRR299 pKa = 11.84PMSEE303 pKa = 3.39ITSFAGLNSGYY314 pKa = 10.5GSEE317 pKa = 4.98VYY319 pKa = 8.8HH320 pKa = 6.71TDD322 pKa = 2.93YY323 pKa = 11.66ALQSKK328 pKa = 9.01QMEE331 pKa = 4.67STTDD335 pKa = 3.58LVDD338 pKa = 3.45VQNANIEE345 pKa = 4.31KK346 pKa = 10.25IMEE349 pKa = 4.16GRR351 pKa = 3.45
Molecular weight: 38.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A160HX24|A0A160HX24_9VIRU Replication associated protein OS=Bovine faeces associated smacovirus 4 OX=1843752 PE=4 SV=1
MM1 pKa = 7.88TIPQSEE7 pKa = 4.21MSKK10 pKa = 10.73RR11 pKa = 11.84EE12 pKa = 3.74LLHH15 pKa = 5.25YY16 pKa = 10.16LRR18 pKa = 11.84HH19 pKa = 6.31RR20 pKa = 11.84SKK22 pKa = 9.59RR23 pKa = 11.84WQFGRR28 pKa = 11.84EE29 pKa = 3.67TGRR32 pKa = 11.84NTGYY36 pKa = 9.93RR37 pKa = 11.84HH38 pKa = 4.48YY39 pKa = 10.24QVRR42 pKa = 11.84YY43 pKa = 8.78EE44 pKa = 4.41SSNDD48 pKa = 3.52DD49 pKa = 3.0IGRR52 pKa = 11.84EE53 pKa = 3.46RR54 pKa = 11.84LYY56 pKa = 10.3WVGIACEE63 pKa = 4.46LEE65 pKa = 4.33PSNGWSNYY73 pKa = 6.52EE74 pKa = 3.99LKK76 pKa = 11.05DD77 pKa = 3.36GDD79 pKa = 5.1FYY81 pKa = 11.27TSDD84 pKa = 3.68DD85 pKa = 4.0ADD87 pKa = 3.67LGKK90 pKa = 10.72YY91 pKa = 9.54RR92 pKa = 11.84FGKK95 pKa = 9.59LRR97 pKa = 11.84KK98 pKa = 8.51FQTRR102 pKa = 11.84LLEE105 pKa = 4.1YY106 pKa = 10.72LEE108 pKa = 4.43SRR110 pKa = 11.84NDD112 pKa = 3.29RR113 pKa = 11.84TVGIVVDD120 pKa = 3.78HH121 pKa = 6.5VGGKK125 pKa = 9.92GKK127 pKa = 8.97TYY129 pKa = 9.82FARR132 pKa = 11.84YY133 pKa = 9.13CSLNHH138 pKa = 6.4KK139 pKa = 9.91AVYY142 pKa = 10.03LHH144 pKa = 7.31GSGTDD149 pKa = 3.36NQVARR154 pKa = 11.84DD155 pKa = 3.98LYY157 pKa = 10.96DD158 pKa = 3.22ISEE161 pKa = 4.33SEE163 pKa = 4.69RR164 pKa = 11.84INAVIIDD171 pKa = 4.01CTRR174 pKa = 11.84SEE176 pKa = 4.33NNLSEE181 pKa = 4.2SDD183 pKa = 3.47FWNGIEE189 pKa = 4.38QIKK192 pKa = 10.6NGHH195 pKa = 5.94LCDD198 pKa = 3.1RR199 pKa = 11.84RR200 pKa = 11.84YY201 pKa = 10.64GYY203 pKa = 9.8RR204 pKa = 11.84EE205 pKa = 3.32KK206 pKa = 10.26WIRR209 pKa = 11.84PPAVLILSNHH219 pKa = 6.09DD220 pKa = 4.3PDD222 pKa = 4.04WKK224 pKa = 10.7CLSEE228 pKa = 4.58DD229 pKa = 3.02RR230 pKa = 11.84WHH232 pKa = 6.99KK233 pKa = 10.48FFIKK237 pKa = 9.67EE238 pKa = 3.71DD239 pKa = 3.32AMYY242 pKa = 10.32EE243 pKa = 4.03YY244 pKa = 11.03YY245 pKa = 10.43KK246 pKa = 10.45RR247 pKa = 11.84EE248 pKa = 4.02KK249 pKa = 10.64KK250 pKa = 8.95VTSLL254 pKa = 3.64
MM1 pKa = 7.88TIPQSEE7 pKa = 4.21MSKK10 pKa = 10.73RR11 pKa = 11.84EE12 pKa = 3.74LLHH15 pKa = 5.25YY16 pKa = 10.16LRR18 pKa = 11.84HH19 pKa = 6.31RR20 pKa = 11.84SKK22 pKa = 9.59RR23 pKa = 11.84WQFGRR28 pKa = 11.84EE29 pKa = 3.67TGRR32 pKa = 11.84NTGYY36 pKa = 9.93RR37 pKa = 11.84HH38 pKa = 4.48YY39 pKa = 10.24QVRR42 pKa = 11.84YY43 pKa = 8.78EE44 pKa = 4.41SSNDD48 pKa = 3.52DD49 pKa = 3.0IGRR52 pKa = 11.84EE53 pKa = 3.46RR54 pKa = 11.84LYY56 pKa = 10.3WVGIACEE63 pKa = 4.46LEE65 pKa = 4.33PSNGWSNYY73 pKa = 6.52EE74 pKa = 3.99LKK76 pKa = 11.05DD77 pKa = 3.36GDD79 pKa = 5.1FYY81 pKa = 11.27TSDD84 pKa = 3.68DD85 pKa = 4.0ADD87 pKa = 3.67LGKK90 pKa = 10.72YY91 pKa = 9.54RR92 pKa = 11.84FGKK95 pKa = 9.59LRR97 pKa = 11.84KK98 pKa = 8.51FQTRR102 pKa = 11.84LLEE105 pKa = 4.1YY106 pKa = 10.72LEE108 pKa = 4.43SRR110 pKa = 11.84NDD112 pKa = 3.29RR113 pKa = 11.84TVGIVVDD120 pKa = 3.78HH121 pKa = 6.5VGGKK125 pKa = 9.92GKK127 pKa = 8.97TYY129 pKa = 9.82FARR132 pKa = 11.84YY133 pKa = 9.13CSLNHH138 pKa = 6.4KK139 pKa = 9.91AVYY142 pKa = 10.03LHH144 pKa = 7.31GSGTDD149 pKa = 3.36NQVARR154 pKa = 11.84DD155 pKa = 3.98LYY157 pKa = 10.96DD158 pKa = 3.22ISEE161 pKa = 4.33SEE163 pKa = 4.69RR164 pKa = 11.84INAVIIDD171 pKa = 4.01CTRR174 pKa = 11.84SEE176 pKa = 4.33NNLSEE181 pKa = 4.2SDD183 pKa = 3.47FWNGIEE189 pKa = 4.38QIKK192 pKa = 10.6NGHH195 pKa = 5.94LCDD198 pKa = 3.1RR199 pKa = 11.84RR200 pKa = 11.84YY201 pKa = 10.64GYY203 pKa = 9.8RR204 pKa = 11.84EE205 pKa = 3.32KK206 pKa = 10.26WIRR209 pKa = 11.84PPAVLILSNHH219 pKa = 6.09DD220 pKa = 4.3PDD222 pKa = 4.04WKK224 pKa = 10.7CLSEE228 pKa = 4.58DD229 pKa = 3.02RR230 pKa = 11.84WHH232 pKa = 6.99KK233 pKa = 10.48FFIKK237 pKa = 9.67EE238 pKa = 3.71DD239 pKa = 3.32AMYY242 pKa = 10.32EE243 pKa = 4.03YY244 pKa = 11.03YY245 pKa = 10.43KK246 pKa = 10.45RR247 pKa = 11.84EE248 pKa = 4.02KK249 pKa = 10.64KK250 pKa = 8.95VTSLL254 pKa = 3.64
Molecular weight: 30.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
605 |
254 |
351 |
302.5 |
34.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.628 ± 0.935 | 0.992 ± 0.618 |
6.777 ± 0.445 | 5.289 ± 1.635 |
2.81 ± 0.215 | 7.438 ± 0.222 |
2.314 ± 0.778 | 5.289 ± 0.108 |
4.628 ± 1.306 | 7.934 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.636 ± 1.553 | 5.289 ± 0.108 |
4.132 ± 1.369 | 3.471 ± 0.702 |
6.612 ± 2.044 | 9.091 ± 1.019 |
5.62 ± 1.065 | 7.273 ± 1.861 |
1.157 ± 1.012 | 5.62 ± 0.928 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
