
Human papillomavirus 184
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 25
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
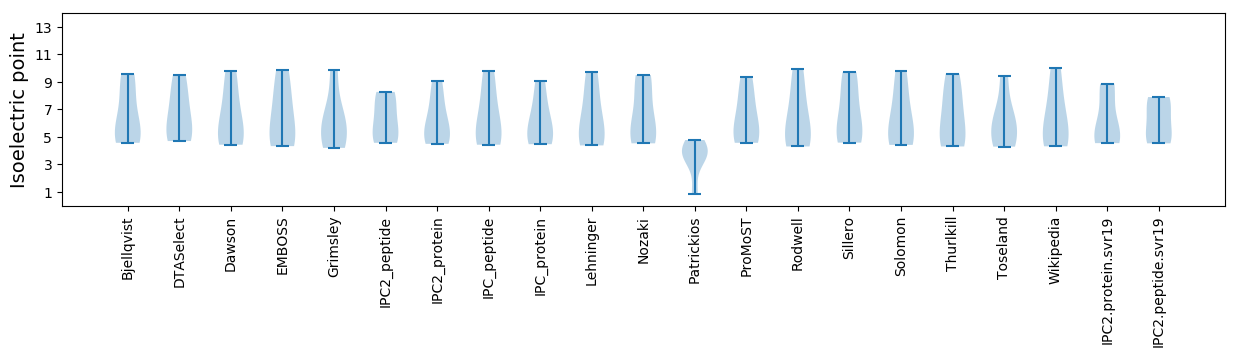
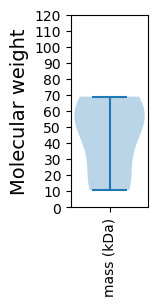
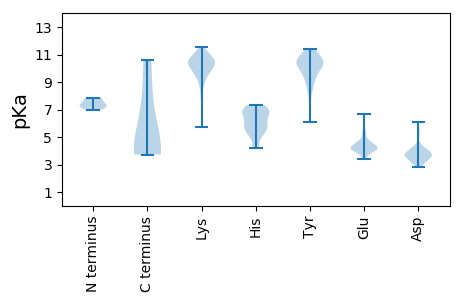
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U6EMG5|U6EMG5_9PAPI Protein E6 OS=Human papillomavirus 184 OX=1472343 GN=E6 PE=3 SV=1
MM1 pKa = 7.3YY2 pKa = 10.34KK3 pKa = 9.86RR4 pKa = 11.84AKK6 pKa = 9.53RR7 pKa = 11.84KK8 pKa = 10.03KK9 pKa = 9.58RR10 pKa = 11.84DD11 pKa = 3.59SVDD14 pKa = 2.89NLYY17 pKa = 10.62RR18 pKa = 11.84KK19 pKa = 9.64CALGADD25 pKa = 4.31CPPDD29 pKa = 3.37VRR31 pKa = 11.84NKK33 pKa = 10.19VEE35 pKa = 4.02NTTLADD41 pKa = 3.06ILLQIFGSAVYY52 pKa = 10.24LGNLGIGTGKK62 pKa = 8.8GTGGTTGYY70 pKa = 10.82SSLAPSITRR79 pKa = 11.84PLPKK83 pKa = 8.64PTRR86 pKa = 11.84PTRR89 pKa = 11.84PFNIPLDD96 pKa = 4.51PIGGGFGSRR105 pKa = 11.84PIGQEE110 pKa = 3.76VPTDD114 pKa = 3.64IIDD117 pKa = 4.12PLSSSIVPLQEE128 pKa = 3.93EE129 pKa = 4.3VGEE132 pKa = 4.26TNIVITADD140 pKa = 3.49PGTSLGGGEE149 pKa = 4.6VVTSTTTVLNPAYY162 pKa = 9.37EE163 pKa = 4.4SEE165 pKa = 4.06GLPDD169 pKa = 3.37VFSRR173 pKa = 11.84GDD175 pKa = 3.28SSIAVLDD182 pKa = 4.48VIPSEE187 pKa = 4.33PPLKK191 pKa = 10.3KK192 pKa = 10.47VAIDD196 pKa = 4.08LPAPSDD202 pKa = 3.73VEE204 pKa = 4.54NVITTLTSVGSDD216 pKa = 2.88FNVYY220 pKa = 9.97VDD222 pKa = 3.88PLEE225 pKa = 4.46TGDD228 pKa = 5.15LIGLEE233 pKa = 4.43EE234 pKa = 5.48IEE236 pKa = 4.58LQPIPKK242 pKa = 10.03YY243 pKa = 9.29STFDD247 pKa = 3.18IEE249 pKa = 4.69EE250 pKa = 4.61PPSTSTPANTVQRR263 pKa = 11.84YY264 pKa = 8.45AGKK267 pKa = 10.43AKK269 pKa = 10.29SLYY272 pKa = 10.3SKK274 pKa = 10.28YY275 pKa = 10.13IKK277 pKa = 9.95QLQTRR282 pKa = 11.84NPDD285 pKa = 3.53FVRR288 pKa = 11.84QPSRR292 pKa = 11.84AVQFEE297 pKa = 4.23FEE299 pKa = 4.7NPAFDD304 pKa = 6.11DD305 pKa = 5.08DD306 pKa = 5.48ISLQFQQDD314 pKa = 3.61LDD316 pKa = 4.08EE317 pKa = 4.63VAAAPDD323 pKa = 3.48IDD325 pKa = 4.2FQDD328 pKa = 2.93VRR330 pKa = 11.84YY331 pKa = 10.04LGRR334 pKa = 11.84PQFLTTEE341 pKa = 4.17SGTVRR346 pKa = 11.84LSRR349 pKa = 11.84LGSKK353 pKa = 9.05ATVTTRR359 pKa = 11.84SGLTVGQKK367 pKa = 8.01VHH369 pKa = 6.95FYY371 pKa = 10.77YY372 pKa = 10.71DD373 pKa = 3.66FSTIYY378 pKa = 10.13PEE380 pKa = 4.68QSFEE384 pKa = 4.04LQPIGEE390 pKa = 4.16FSGLNTYY397 pKa = 9.41VNPLQEE403 pKa = 4.2STFFDD408 pKa = 3.6SSFAHH413 pKa = 6.66ADD415 pKa = 3.08VGEE418 pKa = 4.19IEE420 pKa = 4.4FPEE423 pKa = 4.43VDD425 pKa = 4.49LEE427 pKa = 5.4DD428 pKa = 4.1NMLEE432 pKa = 4.2NFDD435 pKa = 4.09NAHH438 pKa = 6.91LIITGITDD446 pKa = 3.18EE447 pKa = 4.49NEE449 pKa = 3.81QLFIPTLASNIPLKK463 pKa = 10.73PFIPYY468 pKa = 9.57IHH470 pKa = 7.0FGNSTDD476 pKa = 3.88RR477 pKa = 11.84PIVPDD482 pKa = 3.68DD483 pKa = 4.17TIISPTYY490 pKa = 9.55PYY492 pKa = 10.61TPLEE496 pKa = 4.11PANAIEE502 pKa = 4.2VYY504 pKa = 10.96YY505 pKa = 10.91DD506 pKa = 3.94FFLHH510 pKa = 7.13PALQKK515 pKa = 10.16RR516 pKa = 11.84KK517 pKa = 9.36RR518 pKa = 11.84RR519 pKa = 11.84RR520 pKa = 11.84LDD522 pKa = 3.06VFF524 pKa = 3.78
MM1 pKa = 7.3YY2 pKa = 10.34KK3 pKa = 9.86RR4 pKa = 11.84AKK6 pKa = 9.53RR7 pKa = 11.84KK8 pKa = 10.03KK9 pKa = 9.58RR10 pKa = 11.84DD11 pKa = 3.59SVDD14 pKa = 2.89NLYY17 pKa = 10.62RR18 pKa = 11.84KK19 pKa = 9.64CALGADD25 pKa = 4.31CPPDD29 pKa = 3.37VRR31 pKa = 11.84NKK33 pKa = 10.19VEE35 pKa = 4.02NTTLADD41 pKa = 3.06ILLQIFGSAVYY52 pKa = 10.24LGNLGIGTGKK62 pKa = 8.8GTGGTTGYY70 pKa = 10.82SSLAPSITRR79 pKa = 11.84PLPKK83 pKa = 8.64PTRR86 pKa = 11.84PTRR89 pKa = 11.84PFNIPLDD96 pKa = 4.51PIGGGFGSRR105 pKa = 11.84PIGQEE110 pKa = 3.76VPTDD114 pKa = 3.64IIDD117 pKa = 4.12PLSSSIVPLQEE128 pKa = 3.93EE129 pKa = 4.3VGEE132 pKa = 4.26TNIVITADD140 pKa = 3.49PGTSLGGGEE149 pKa = 4.6VVTSTTTVLNPAYY162 pKa = 9.37EE163 pKa = 4.4SEE165 pKa = 4.06GLPDD169 pKa = 3.37VFSRR173 pKa = 11.84GDD175 pKa = 3.28SSIAVLDD182 pKa = 4.48VIPSEE187 pKa = 4.33PPLKK191 pKa = 10.3KK192 pKa = 10.47VAIDD196 pKa = 4.08LPAPSDD202 pKa = 3.73VEE204 pKa = 4.54NVITTLTSVGSDD216 pKa = 2.88FNVYY220 pKa = 9.97VDD222 pKa = 3.88PLEE225 pKa = 4.46TGDD228 pKa = 5.15LIGLEE233 pKa = 4.43EE234 pKa = 5.48IEE236 pKa = 4.58LQPIPKK242 pKa = 10.03YY243 pKa = 9.29STFDD247 pKa = 3.18IEE249 pKa = 4.69EE250 pKa = 4.61PPSTSTPANTVQRR263 pKa = 11.84YY264 pKa = 8.45AGKK267 pKa = 10.43AKK269 pKa = 10.29SLYY272 pKa = 10.3SKK274 pKa = 10.28YY275 pKa = 10.13IKK277 pKa = 9.95QLQTRR282 pKa = 11.84NPDD285 pKa = 3.53FVRR288 pKa = 11.84QPSRR292 pKa = 11.84AVQFEE297 pKa = 4.23FEE299 pKa = 4.7NPAFDD304 pKa = 6.11DD305 pKa = 5.08DD306 pKa = 5.48ISLQFQQDD314 pKa = 3.61LDD316 pKa = 4.08EE317 pKa = 4.63VAAAPDD323 pKa = 3.48IDD325 pKa = 4.2FQDD328 pKa = 2.93VRR330 pKa = 11.84YY331 pKa = 10.04LGRR334 pKa = 11.84PQFLTTEE341 pKa = 4.17SGTVRR346 pKa = 11.84LSRR349 pKa = 11.84LGSKK353 pKa = 9.05ATVTTRR359 pKa = 11.84SGLTVGQKK367 pKa = 8.01VHH369 pKa = 6.95FYY371 pKa = 10.77YY372 pKa = 10.71DD373 pKa = 3.66FSTIYY378 pKa = 10.13PEE380 pKa = 4.68QSFEE384 pKa = 4.04LQPIGEE390 pKa = 4.16FSGLNTYY397 pKa = 9.41VNPLQEE403 pKa = 4.2STFFDD408 pKa = 3.6SSFAHH413 pKa = 6.66ADD415 pKa = 3.08VGEE418 pKa = 4.19IEE420 pKa = 4.4FPEE423 pKa = 4.43VDD425 pKa = 4.49LEE427 pKa = 5.4DD428 pKa = 4.1NMLEE432 pKa = 4.2NFDD435 pKa = 4.09NAHH438 pKa = 6.91LIITGITDD446 pKa = 3.18EE447 pKa = 4.49NEE449 pKa = 3.81QLFIPTLASNIPLKK463 pKa = 10.73PFIPYY468 pKa = 9.57IHH470 pKa = 7.0FGNSTDD476 pKa = 3.88RR477 pKa = 11.84PIVPDD482 pKa = 3.68DD483 pKa = 4.17TIISPTYY490 pKa = 9.55PYY492 pKa = 10.61TPLEE496 pKa = 4.11PANAIEE502 pKa = 4.2VYY504 pKa = 10.96YY505 pKa = 10.91DD506 pKa = 3.94FFLHH510 pKa = 7.13PALQKK515 pKa = 10.16RR516 pKa = 11.84KK517 pKa = 9.36RR518 pKa = 11.84RR519 pKa = 11.84RR520 pKa = 11.84LDD522 pKa = 3.06VFF524 pKa = 3.78
Molecular weight: 57.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U6ELR9|U6ELR9_9PAPI Minor capsid protein L2 OS=Human papillomavirus 184 OX=1472343 GN=L2 PE=3 SV=1
MM1 pKa = 7.0NQADD5 pKa = 3.68LTARR9 pKa = 11.84FDD11 pKa = 3.89ALQDD15 pKa = 3.63TLMTLYY21 pKa = 10.16EE22 pKa = 4.33RR23 pKa = 11.84NATDD27 pKa = 3.18IDD29 pKa = 3.99SQILHH34 pKa = 6.73WEE36 pKa = 4.49TVRR39 pKa = 11.84KK40 pKa = 9.93QYY42 pKa = 9.59VTMYY46 pKa = 8.64YY47 pKa = 10.46ARR49 pKa = 11.84KK50 pKa = 9.03EE51 pKa = 4.47GYY53 pKa = 9.35RR54 pKa = 11.84SLGMQPLPALTVSEE68 pKa = 4.61YY69 pKa = 10.1KK70 pKa = 10.68AKK72 pKa = 10.46EE73 pKa = 4.13AIQMVILLRR82 pKa = 11.84SLRR85 pKa = 11.84QSQFSNEE92 pKa = 3.85LWTLTDD98 pKa = 3.58TSAEE102 pKa = 4.07LLHH105 pKa = 5.96TQPKK109 pKa = 10.13NCFKK113 pKa = 10.62KK114 pKa = 9.62HH115 pKa = 5.14GYY117 pKa = 6.11TVEE120 pKa = 4.13VYY122 pKa = 10.56FDD124 pKa = 3.94EE125 pKa = 5.18NPANVFPYY133 pKa = 9.96TNWDD137 pKa = 3.45SIYY140 pKa = 10.4YY141 pKa = 9.41QDD143 pKa = 5.58SHH145 pKa = 5.73EE146 pKa = 4.27QWHH149 pKa = 5.67KK150 pKa = 9.01TAGLVDD156 pKa = 3.78YY157 pKa = 10.94DD158 pKa = 3.56GMYY161 pKa = 10.43FQEE164 pKa = 4.52TNGDD168 pKa = 3.03RR169 pKa = 11.84TYY171 pKa = 11.18FQLFHH176 pKa = 7.28KK177 pKa = 10.64DD178 pKa = 2.85ALRR181 pKa = 11.84YY182 pKa = 9.84GSTGRR187 pKa = 11.84WSVHH191 pKa = 5.37YY192 pKa = 10.0KK193 pKa = 9.97NQTILPSASVSSSTRR208 pKa = 11.84QPVDD212 pKa = 3.45EE213 pKa = 4.59PAEE216 pKa = 4.26TTAYY220 pKa = 9.8PSTPPQIAAGGSQRR234 pKa = 11.84SQTEE238 pKa = 3.53EE239 pKa = 4.01RR240 pKa = 11.84GTVSSTQQTPPATISSVRR258 pKa = 11.84LRR260 pKa = 11.84RR261 pKa = 11.84RR262 pKa = 11.84RR263 pKa = 11.84GQGEE267 pKa = 3.79RR268 pKa = 11.84TTRR271 pKa = 11.84EE272 pKa = 3.72RR273 pKa = 11.84EE274 pKa = 4.12SPTKK278 pKa = 10.16RR279 pKa = 11.84RR280 pKa = 11.84RR281 pKa = 11.84TTEE284 pKa = 3.53TDD286 pKa = 3.01SARR289 pKa = 11.84LEE291 pKa = 4.33SVPAPEE297 pKa = 4.78AVGTRR302 pKa = 11.84HH303 pKa = 5.32RR304 pKa = 11.84TVPSTGLSRR313 pKa = 11.84LGRR316 pKa = 11.84LQEE319 pKa = 4.04EE320 pKa = 4.47ARR322 pKa = 11.84DD323 pKa = 3.79PPAILVTGPGNSLKK337 pKa = 10.28CWRR340 pKa = 11.84WRR342 pKa = 11.84LKK344 pKa = 10.64KK345 pKa = 10.56FSRR348 pKa = 11.84YY349 pKa = 6.52YY350 pKa = 11.26LRR352 pKa = 11.84MSTVWSWAGEE362 pKa = 4.25VCPKK366 pKa = 10.26ASKK369 pKa = 10.84NRR371 pKa = 11.84MLIAFRR377 pKa = 11.84DD378 pKa = 3.66NEE380 pKa = 4.01QRR382 pKa = 11.84SVFLKK387 pKa = 10.22LVSMPKK393 pKa = 8.51HH394 pKa = 4.54TTFGLAQLGILL405 pKa = 4.22
MM1 pKa = 7.0NQADD5 pKa = 3.68LTARR9 pKa = 11.84FDD11 pKa = 3.89ALQDD15 pKa = 3.63TLMTLYY21 pKa = 10.16EE22 pKa = 4.33RR23 pKa = 11.84NATDD27 pKa = 3.18IDD29 pKa = 3.99SQILHH34 pKa = 6.73WEE36 pKa = 4.49TVRR39 pKa = 11.84KK40 pKa = 9.93QYY42 pKa = 9.59VTMYY46 pKa = 8.64YY47 pKa = 10.46ARR49 pKa = 11.84KK50 pKa = 9.03EE51 pKa = 4.47GYY53 pKa = 9.35RR54 pKa = 11.84SLGMQPLPALTVSEE68 pKa = 4.61YY69 pKa = 10.1KK70 pKa = 10.68AKK72 pKa = 10.46EE73 pKa = 4.13AIQMVILLRR82 pKa = 11.84SLRR85 pKa = 11.84QSQFSNEE92 pKa = 3.85LWTLTDD98 pKa = 3.58TSAEE102 pKa = 4.07LLHH105 pKa = 5.96TQPKK109 pKa = 10.13NCFKK113 pKa = 10.62KK114 pKa = 9.62HH115 pKa = 5.14GYY117 pKa = 6.11TVEE120 pKa = 4.13VYY122 pKa = 10.56FDD124 pKa = 3.94EE125 pKa = 5.18NPANVFPYY133 pKa = 9.96TNWDD137 pKa = 3.45SIYY140 pKa = 10.4YY141 pKa = 9.41QDD143 pKa = 5.58SHH145 pKa = 5.73EE146 pKa = 4.27QWHH149 pKa = 5.67KK150 pKa = 9.01TAGLVDD156 pKa = 3.78YY157 pKa = 10.94DD158 pKa = 3.56GMYY161 pKa = 10.43FQEE164 pKa = 4.52TNGDD168 pKa = 3.03RR169 pKa = 11.84TYY171 pKa = 11.18FQLFHH176 pKa = 7.28KK177 pKa = 10.64DD178 pKa = 2.85ALRR181 pKa = 11.84YY182 pKa = 9.84GSTGRR187 pKa = 11.84WSVHH191 pKa = 5.37YY192 pKa = 10.0KK193 pKa = 9.97NQTILPSASVSSSTRR208 pKa = 11.84QPVDD212 pKa = 3.45EE213 pKa = 4.59PAEE216 pKa = 4.26TTAYY220 pKa = 9.8PSTPPQIAAGGSQRR234 pKa = 11.84SQTEE238 pKa = 3.53EE239 pKa = 4.01RR240 pKa = 11.84GTVSSTQQTPPATISSVRR258 pKa = 11.84LRR260 pKa = 11.84RR261 pKa = 11.84RR262 pKa = 11.84RR263 pKa = 11.84GQGEE267 pKa = 3.79RR268 pKa = 11.84TTRR271 pKa = 11.84EE272 pKa = 3.72RR273 pKa = 11.84EE274 pKa = 4.12SPTKK278 pKa = 10.16RR279 pKa = 11.84RR280 pKa = 11.84RR281 pKa = 11.84TTEE284 pKa = 3.53TDD286 pKa = 3.01SARR289 pKa = 11.84LEE291 pKa = 4.33SVPAPEE297 pKa = 4.78AVGTRR302 pKa = 11.84HH303 pKa = 5.32RR304 pKa = 11.84TVPSTGLSRR313 pKa = 11.84LGRR316 pKa = 11.84LQEE319 pKa = 4.04EE320 pKa = 4.47ARR322 pKa = 11.84DD323 pKa = 3.79PPAILVTGPGNSLKK337 pKa = 10.28CWRR340 pKa = 11.84WRR342 pKa = 11.84LKK344 pKa = 10.64KK345 pKa = 10.56FSRR348 pKa = 11.84YY349 pKa = 6.52YY350 pKa = 11.26LRR352 pKa = 11.84MSTVWSWAGEE362 pKa = 4.25VCPKK366 pKa = 10.26ASKK369 pKa = 10.84NRR371 pKa = 11.84MLIAFRR377 pKa = 11.84DD378 pKa = 3.66NEE380 pKa = 4.01QRR382 pKa = 11.84SVFLKK387 pKa = 10.22LVSMPKK393 pKa = 8.51HH394 pKa = 4.54TTFGLAQLGILL405 pKa = 4.22
Molecular weight: 46.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2293 |
97 |
603 |
382.2 |
43.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.582 ± 0.565 | 2.268 ± 0.809 |
6.28 ± 0.468 | 6.149 ± 0.439 |
4.797 ± 0.392 | 5.451 ± 0.83 |
1.57 ± 0.194 | 4.841 ± 0.624 |
5.539 ± 0.653 | 9.202 ± 0.517 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.875 ± 0.448 | 4.928 ± 0.673 |
5.931 ± 1.139 | 4.361 ± 0.326 |
5.408 ± 0.688 | 7.327 ± 0.836 |
6.847 ± 0.728 | 6.149 ± 0.336 |
1.221 ± 0.316 | 4.274 ± 0.5 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
