
Piptocephalis cylindrospora
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Zoopagomycota; Zoopagomycotina; Zoopagomycetes; Zoopagales; Piptocephalidaceae; Piptocephalis
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
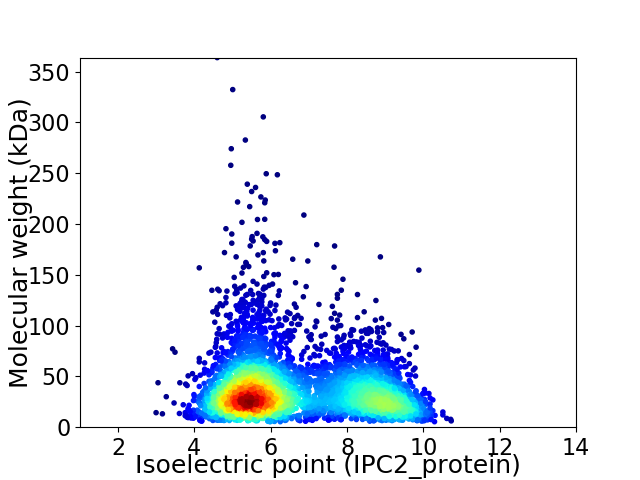
Virtual 2D-PAGE plot for 4271 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P9Y3C8|A0A4P9Y3C8_9FUNG BZIP domain-containing protein OS=Piptocephalis cylindrospora OX=1907219 GN=BJ684DRAFT_20301 PE=4 SV=1
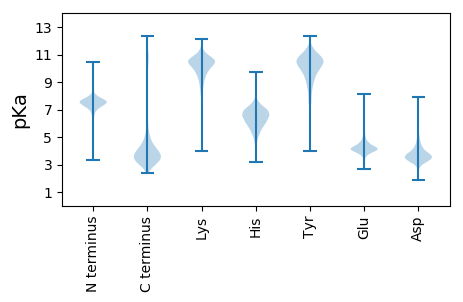
MM1 pKa = 7.51KK2 pKa = 10.36SALILAAAAAAITPASATLFSSTLACIVNSQRR34 pKa = 11.84QKK36 pKa = 11.27YY37 pKa = 10.15DD38 pKa = 3.24LTPVKK43 pKa = 8.65TTTADD48 pKa = 3.45FNEE51 pKa = 4.73LQTEE55 pKa = 4.56AHH57 pKa = 5.89QMSSTKK63 pKa = 10.33KK64 pKa = 9.81VIPGSFGNGWRR75 pKa = 11.84KK76 pKa = 9.63IPVQFTCPYY85 pKa = 7.79AHH87 pKa = 7.39EE88 pKa = 4.26YY89 pKa = 11.08DD90 pKa = 3.68GTNEE94 pKa = 3.77VSVVNVINNVTHH106 pKa = 7.36SMDD109 pKa = 3.03ILNAKK114 pKa = 9.91NITAVSCSAHH124 pKa = 6.49KK125 pKa = 10.57EE126 pKa = 3.53VDD128 pKa = 3.59QVDD131 pKa = 3.69KK132 pKa = 10.67KK133 pKa = 9.42TGKK136 pKa = 8.41KK137 pKa = 7.9TRR139 pKa = 11.84NSHH142 pKa = 4.85CWGMVKK148 pKa = 9.54IYY150 pKa = 9.57YY151 pKa = 8.89TDD153 pKa = 2.97SGNFVPAANNCTSTPPPTDD172 pKa = 2.82GGYY175 pKa = 8.38TPPGDD180 pKa = 3.9GNQDD184 pKa = 2.89TCYY187 pKa = 10.44EE188 pKa = 4.13GSTDD192 pKa = 4.28PKK194 pKa = 9.28CTSPGGDD201 pKa = 3.09TTYY204 pKa = 10.48TPPGGDD210 pKa = 2.99TTYY213 pKa = 10.58TPPGDD218 pKa = 3.79DD219 pKa = 3.06TTYY222 pKa = 10.84TPPGDD227 pKa = 3.9NNQDD231 pKa = 2.95TCYY234 pKa = 10.51EE235 pKa = 3.99GSTDD239 pKa = 4.43SKK241 pKa = 10.01CTSPGGDD248 pKa = 3.17TTYY251 pKa = 10.71TPPDD255 pKa = 3.51NKK257 pKa = 10.57DD258 pKa = 4.04DD259 pKa = 4.15EE260 pKa = 4.89SCDD263 pKa = 4.34DD264 pKa = 3.68STTPGNGGTSPGGDD278 pKa = 3.04TTYY281 pKa = 10.73TPPGDD286 pKa = 3.58NTTYY290 pKa = 10.36TPPGKK295 pKa = 10.11DD296 pKa = 3.78DD297 pKa = 5.29DD298 pKa = 4.68EE299 pKa = 6.22KK300 pKa = 11.47CDD302 pKa = 3.82IYY304 pKa = 10.93STSDD308 pKa = 2.61KK309 pKa = 10.8CNPGNEE315 pKa = 4.33DD316 pKa = 4.04EE317 pKa = 6.05GDD319 pKa = 3.68DD320 pKa = 4.05TYY322 pKa = 11.59TPPGKK327 pKa = 10.14DD328 pKa = 3.78DD329 pKa = 5.29DD330 pKa = 4.68EE331 pKa = 6.22KK332 pKa = 11.47CDD334 pKa = 3.82IYY336 pKa = 10.93STSDD340 pKa = 2.71KK341 pKa = 10.98CNPGNDD347 pKa = 4.19KK348 pKa = 11.39DD349 pKa = 3.92DD350 pKa = 3.69TTYY353 pKa = 11.71SNDD356 pKa = 3.67EE357 pKa = 4.5DD358 pKa = 3.84DD359 pKa = 3.71TTYY362 pKa = 11.66SNDD365 pKa = 3.67EE366 pKa = 4.5DD367 pKa = 3.84DD368 pKa = 3.71TTYY371 pKa = 11.66SNDD374 pKa = 3.54EE375 pKa = 4.77DD376 pKa = 5.9DD377 pKa = 5.82SGDD380 pKa = 4.14DD381 pKa = 4.41SDD383 pKa = 6.28DD384 pKa = 4.61GSDD387 pKa = 3.97DD388 pKa = 4.13DD389 pKa = 6.24QYY391 pKa = 12.14NQDD394 pKa = 3.17NGYY397 pKa = 10.59
MM1 pKa = 7.51KK2 pKa = 10.36SALILAAAAAAITPASATLFSSTLACIVNSQRR34 pKa = 11.84QKK36 pKa = 11.27YY37 pKa = 10.15DD38 pKa = 3.24LTPVKK43 pKa = 8.65TTTADD48 pKa = 3.45FNEE51 pKa = 4.73LQTEE55 pKa = 4.56AHH57 pKa = 5.89QMSSTKK63 pKa = 10.33KK64 pKa = 9.81VIPGSFGNGWRR75 pKa = 11.84KK76 pKa = 9.63IPVQFTCPYY85 pKa = 7.79AHH87 pKa = 7.39EE88 pKa = 4.26YY89 pKa = 11.08DD90 pKa = 3.68GTNEE94 pKa = 3.77VSVVNVINNVTHH106 pKa = 7.36SMDD109 pKa = 3.03ILNAKK114 pKa = 9.91NITAVSCSAHH124 pKa = 6.49KK125 pKa = 10.57EE126 pKa = 3.53VDD128 pKa = 3.59QVDD131 pKa = 3.69KK132 pKa = 10.67KK133 pKa = 9.42TGKK136 pKa = 8.41KK137 pKa = 7.9TRR139 pKa = 11.84NSHH142 pKa = 4.85CWGMVKK148 pKa = 9.54IYY150 pKa = 9.57YY151 pKa = 8.89TDD153 pKa = 2.97SGNFVPAANNCTSTPPPTDD172 pKa = 2.82GGYY175 pKa = 8.38TPPGDD180 pKa = 3.9GNQDD184 pKa = 2.89TCYY187 pKa = 10.44EE188 pKa = 4.13GSTDD192 pKa = 4.28PKK194 pKa = 9.28CTSPGGDD201 pKa = 3.09TTYY204 pKa = 10.48TPPGGDD210 pKa = 2.99TTYY213 pKa = 10.58TPPGDD218 pKa = 3.79DD219 pKa = 3.06TTYY222 pKa = 10.84TPPGDD227 pKa = 3.9NNQDD231 pKa = 2.95TCYY234 pKa = 10.51EE235 pKa = 3.99GSTDD239 pKa = 4.43SKK241 pKa = 10.01CTSPGGDD248 pKa = 3.17TTYY251 pKa = 10.71TPPDD255 pKa = 3.51NKK257 pKa = 10.57DD258 pKa = 4.04DD259 pKa = 4.15EE260 pKa = 4.89SCDD263 pKa = 4.34DD264 pKa = 3.68STTPGNGGTSPGGDD278 pKa = 3.04TTYY281 pKa = 10.73TPPGDD286 pKa = 3.58NTTYY290 pKa = 10.36TPPGKK295 pKa = 10.11DD296 pKa = 3.78DD297 pKa = 5.29DD298 pKa = 4.68EE299 pKa = 6.22KK300 pKa = 11.47CDD302 pKa = 3.82IYY304 pKa = 10.93STSDD308 pKa = 2.61KK309 pKa = 10.8CNPGNEE315 pKa = 4.33DD316 pKa = 4.04EE317 pKa = 6.05GDD319 pKa = 3.68DD320 pKa = 4.05TYY322 pKa = 11.59TPPGKK327 pKa = 10.14DD328 pKa = 3.78DD329 pKa = 5.29DD330 pKa = 4.68EE331 pKa = 6.22KK332 pKa = 11.47CDD334 pKa = 3.82IYY336 pKa = 10.93STSDD340 pKa = 2.71KK341 pKa = 10.98CNPGNDD347 pKa = 4.19KK348 pKa = 11.39DD349 pKa = 3.92DD350 pKa = 3.69TTYY353 pKa = 11.71SNDD356 pKa = 3.67EE357 pKa = 4.5DD358 pKa = 3.84DD359 pKa = 3.71TTYY362 pKa = 11.66SNDD365 pKa = 3.67EE366 pKa = 4.5DD367 pKa = 3.84DD368 pKa = 3.71TTYY371 pKa = 11.66SNDD374 pKa = 3.54EE375 pKa = 4.77DD376 pKa = 5.9DD377 pKa = 5.82SGDD380 pKa = 4.14DD381 pKa = 4.41SDD383 pKa = 6.28DD384 pKa = 4.61GSDD387 pKa = 3.97DD388 pKa = 4.13DD389 pKa = 6.24QYY391 pKa = 12.14NQDD394 pKa = 3.17NGYY397 pKa = 10.59
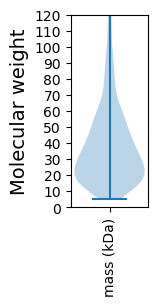
Molecular weight: 42.44 kDa
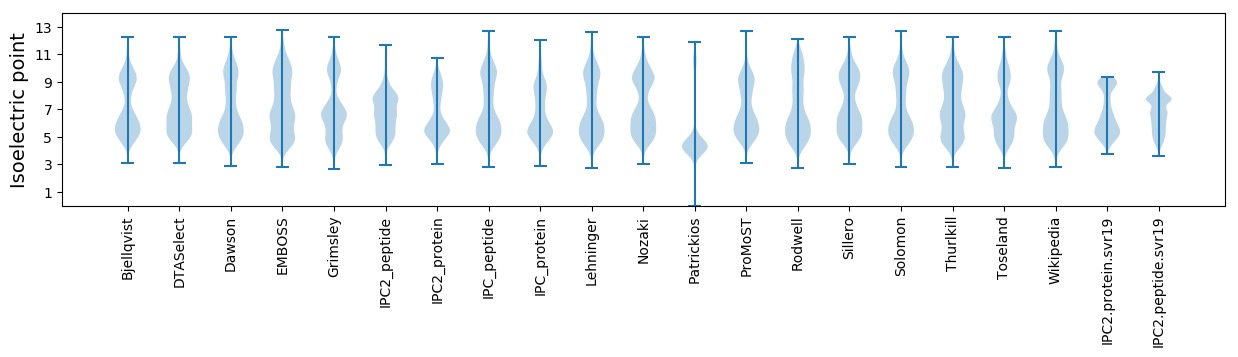
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P9XZC9|A0A4P9XZC9_9FUNG Uncharacterized protein OS=Piptocephalis cylindrospora OX=1907219 GN=BJ684DRAFT_21685 PE=4 SV=1
NN1 pKa = 7.39LEE3 pKa = 4.13LYY5 pKa = 10.13QRR7 pKa = 11.84LNFLHH12 pKa = 6.05QASTLLTLAGPHH24 pKa = 6.26FAPLSRR30 pKa = 11.84SLGRR34 pKa = 11.84DD35 pKa = 2.81MDD37 pKa = 4.43SVRR40 pKa = 11.84KK41 pKa = 9.88RR42 pKa = 11.84STLRR46 pKa = 11.84ADD48 pKa = 3.6PSVKK52 pKa = 9.9RR53 pKa = 11.84QYY55 pKa = 10.98CRR57 pKa = 11.84ACHH60 pKa = 4.82THH62 pKa = 7.3LIEE65 pKa = 4.96GFTLRR70 pKa = 11.84TRR72 pKa = 11.84LTRR75 pKa = 11.84KK76 pKa = 9.18GGKK79 pKa = 9.95AIIRR83 pKa = 11.84TCMYY87 pKa = 10.07CEE89 pKa = 3.83KK90 pKa = 10.37EE91 pKa = 4.5RR92 pKa = 11.84RR93 pKa = 11.84LLCNPQHH100 pKa = 6.97EE101 pKa = 4.7VFNWKK106 pKa = 9.81
NN1 pKa = 7.39LEE3 pKa = 4.13LYY5 pKa = 10.13QRR7 pKa = 11.84LNFLHH12 pKa = 6.05QASTLLTLAGPHH24 pKa = 6.26FAPLSRR30 pKa = 11.84SLGRR34 pKa = 11.84DD35 pKa = 2.81MDD37 pKa = 4.43SVRR40 pKa = 11.84KK41 pKa = 9.88RR42 pKa = 11.84STLRR46 pKa = 11.84ADD48 pKa = 3.6PSVKK52 pKa = 9.9RR53 pKa = 11.84QYY55 pKa = 10.98CRR57 pKa = 11.84ACHH60 pKa = 4.82THH62 pKa = 7.3LIEE65 pKa = 4.96GFTLRR70 pKa = 11.84TRR72 pKa = 11.84LTRR75 pKa = 11.84KK76 pKa = 9.18GGKK79 pKa = 9.95AIIRR83 pKa = 11.84TCMYY87 pKa = 10.07CEE89 pKa = 3.83KK90 pKa = 10.37EE91 pKa = 4.5RR92 pKa = 11.84RR93 pKa = 11.84LLCNPQHH100 pKa = 6.97EE101 pKa = 4.7VFNWKK106 pKa = 9.81
Molecular weight: 12.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1596308 |
50 |
3304 |
373.8 |
41.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.833 ± 0.038 | 1.205 ± 0.014 |
5.39 ± 0.029 | 6.776 ± 0.049 |
3.206 ± 0.027 | 7.599 ± 0.046 |
2.666 ± 0.019 | 4.665 ± 0.028 |
4.502 ± 0.036 | 9.832 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.443 ± 0.016 | 2.696 ± 0.019 |
6.133 ± 0.048 | 3.789 ± 0.022 |
6.718 ± 0.038 | 8.7 ± 0.058 |
5.707 ± 0.029 | 6.044 ± 0.028 |
1.449 ± 0.017 | 2.647 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
