
Actinomyces sp. oral taxon 180 str. F0310
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Actinomycetales; Actinomycetaceae; Actinomyces; unclassified Actinomyces; Actinomyces sp. oral taxon 180
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
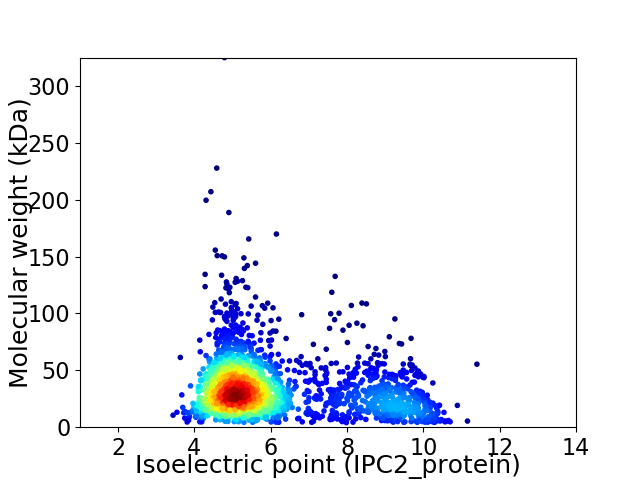
Virtual 2D-PAGE plot for 2070 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E6KRP5|E6KRP5_9ACTO ABC superfamily ATP binding cassette transporter ABC protein OS=Actinomyces sp. oral taxon 180 str. F0310 OX=888052 GN=thrB2 PE=4 SV=1
MM1 pKa = 7.88SIRR4 pKa = 11.84FLRR7 pKa = 11.84SSLRR11 pKa = 11.84AAPLVGATLAVSLLSLAGLAAYY33 pKa = 8.44TGGGEE38 pKa = 4.73AAGQPDD44 pKa = 4.62PSPIVYY50 pKa = 10.11EE51 pKa = 4.65DD52 pKa = 4.35DD53 pKa = 3.92SSLSIVAAFTDD64 pKa = 3.75MTSQLNGSVNTVAYY78 pKa = 8.0QQAYY82 pKa = 10.02RR83 pKa = 11.84GVTYY87 pKa = 10.59DD88 pKa = 3.08KK89 pKa = 10.47EE90 pKa = 4.56AYY92 pKa = 10.09VYY94 pKa = 10.35VPPTYY99 pKa = 11.0SPDD102 pKa = 3.19TPANVVYY109 pKa = 8.88LTHH112 pKa = 6.53GWQGSAAGLAEE123 pKa = 3.91GVVPVVDD130 pKa = 4.2EE131 pKa = 4.36LTASGQLSPTLVVFATYY148 pKa = 10.78YY149 pKa = 9.27PDD151 pKa = 3.68RR152 pKa = 11.84SFAGDD157 pKa = 4.08DD158 pKa = 3.76YY159 pKa = 11.86EE160 pKa = 6.92DD161 pKa = 5.11DD162 pKa = 4.06YY163 pKa = 11.97EE164 pKa = 4.89LNRR167 pKa = 11.84FFATTEE173 pKa = 3.28IDD175 pKa = 3.79TIIDD179 pKa = 3.65AVEE182 pKa = 3.84SRR184 pKa = 11.84YY185 pKa = 8.75ATYY188 pKa = 10.96AGGDD192 pKa = 3.41TSDD195 pKa = 4.57QSLQDD200 pKa = 3.69SRR202 pKa = 11.84THH204 pKa = 6.17RR205 pKa = 11.84AFGGFSMGATTTWDD219 pKa = 3.64VFSMRR224 pKa = 11.84PQYY227 pKa = 10.55FYY229 pKa = 11.05GYY231 pKa = 8.77MPMAGEE237 pKa = 4.04SWIGRR242 pKa = 11.84EE243 pKa = 3.48TDD245 pKa = 3.26ADD247 pKa = 3.44IDD249 pKa = 4.23QIAHH253 pKa = 7.21LIAAGAEE260 pKa = 4.01RR261 pKa = 11.84ADD263 pKa = 3.94YY264 pKa = 10.18GAQDD268 pKa = 3.57FRR270 pKa = 11.84ILASVGSDD278 pKa = 5.34DD279 pKa = 3.97PALWDD284 pKa = 3.7MSPQLEE290 pKa = 4.24QLQLDD295 pKa = 4.66YY296 pKa = 10.73PDD298 pKa = 5.32LMTPGSLQLWIDD310 pKa = 3.5EE311 pKa = 4.83GEE313 pKa = 4.19DD314 pKa = 3.16HH315 pKa = 7.65SMASIQNQVAHH326 pKa = 7.03DD327 pKa = 4.56LPRR330 pKa = 11.84LLPQAA335 pKa = 4.34
MM1 pKa = 7.88SIRR4 pKa = 11.84FLRR7 pKa = 11.84SSLRR11 pKa = 11.84AAPLVGATLAVSLLSLAGLAAYY33 pKa = 8.44TGGGEE38 pKa = 4.73AAGQPDD44 pKa = 4.62PSPIVYY50 pKa = 10.11EE51 pKa = 4.65DD52 pKa = 4.35DD53 pKa = 3.92SSLSIVAAFTDD64 pKa = 3.75MTSQLNGSVNTVAYY78 pKa = 8.0QQAYY82 pKa = 10.02RR83 pKa = 11.84GVTYY87 pKa = 10.59DD88 pKa = 3.08KK89 pKa = 10.47EE90 pKa = 4.56AYY92 pKa = 10.09VYY94 pKa = 10.35VPPTYY99 pKa = 11.0SPDD102 pKa = 3.19TPANVVYY109 pKa = 8.88LTHH112 pKa = 6.53GWQGSAAGLAEE123 pKa = 3.91GVVPVVDD130 pKa = 4.2EE131 pKa = 4.36LTASGQLSPTLVVFATYY148 pKa = 10.78YY149 pKa = 9.27PDD151 pKa = 3.68RR152 pKa = 11.84SFAGDD157 pKa = 4.08DD158 pKa = 3.76YY159 pKa = 11.86EE160 pKa = 6.92DD161 pKa = 5.11DD162 pKa = 4.06YY163 pKa = 11.97EE164 pKa = 4.89LNRR167 pKa = 11.84FFATTEE173 pKa = 3.28IDD175 pKa = 3.79TIIDD179 pKa = 3.65AVEE182 pKa = 3.84SRR184 pKa = 11.84YY185 pKa = 8.75ATYY188 pKa = 10.96AGGDD192 pKa = 3.41TSDD195 pKa = 4.57QSLQDD200 pKa = 3.69SRR202 pKa = 11.84THH204 pKa = 6.17RR205 pKa = 11.84AFGGFSMGATTTWDD219 pKa = 3.64VFSMRR224 pKa = 11.84PQYY227 pKa = 10.55FYY229 pKa = 11.05GYY231 pKa = 8.77MPMAGEE237 pKa = 4.04SWIGRR242 pKa = 11.84EE243 pKa = 3.48TDD245 pKa = 3.26ADD247 pKa = 3.44IDD249 pKa = 4.23QIAHH253 pKa = 7.21LIAAGAEE260 pKa = 4.01RR261 pKa = 11.84ADD263 pKa = 3.94YY264 pKa = 10.18GAQDD268 pKa = 3.57FRR270 pKa = 11.84ILASVGSDD278 pKa = 5.34DD279 pKa = 3.97PALWDD284 pKa = 3.7MSPQLEE290 pKa = 4.24QLQLDD295 pKa = 4.66YY296 pKa = 10.73PDD298 pKa = 5.32LMTPGSLQLWIDD310 pKa = 3.5EE311 pKa = 4.83GEE313 pKa = 4.19DD314 pKa = 3.16HH315 pKa = 7.65SMASIQNQVAHH326 pKa = 7.03DD327 pKa = 4.56LPRR330 pKa = 11.84LLPQAA335 pKa = 4.34
Molecular weight: 36.31 kDa
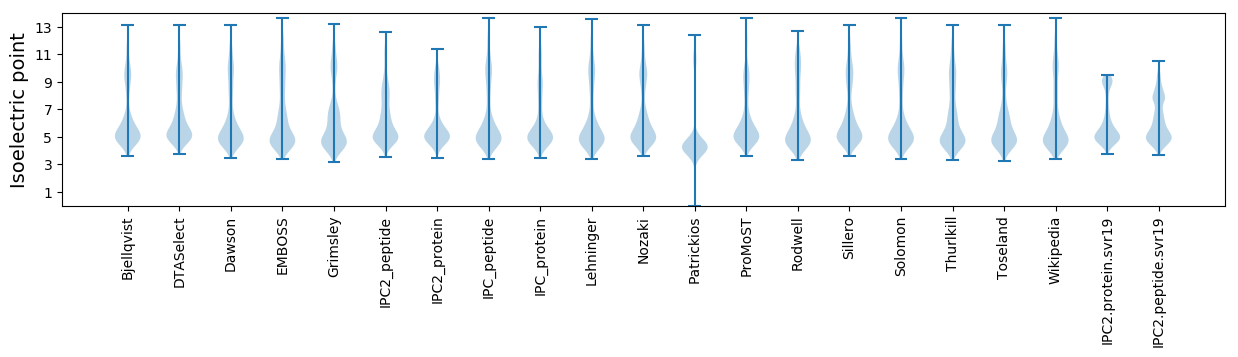
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E6KTB0|E6KTB0_9ACTO Uncharacterized protein (Fragment) OS=Actinomyces sp. oral taxon 180 str. F0310 OX=888052 GN=HMPREF9006_1462 PE=4 SV=1
MM1 pKa = 6.74EE2 pKa = 5.03TAPPTHH8 pKa = 6.95PPLLRR13 pKa = 11.84RR14 pKa = 11.84TSPSKK19 pKa = 10.86KK20 pKa = 8.66PWAAPTSSRR29 pKa = 11.84TVEE32 pKa = 4.23GLQQTRR38 pKa = 11.84PPPTRR43 pKa = 11.84TRR45 pKa = 11.84PQTQGRR51 pKa = 11.84LATAPGTQMAPVRR64 pKa = 11.84LIPTLSRR71 pKa = 11.84GTIPASARR79 pKa = 11.84TLTPRR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84TTPPTVRR93 pKa = 11.84PTTPLIRR100 pKa = 11.84RR101 pKa = 11.84ATATRR106 pKa = 11.84PRR108 pKa = 11.84PRR110 pKa = 11.84KK111 pKa = 8.01TSRR114 pKa = 11.84TSPRR118 pKa = 11.84RR119 pKa = 11.84ISPTRR124 pKa = 11.84KK125 pKa = 5.37TTPTASPIRR134 pKa = 11.84TRR136 pKa = 11.84PTRR139 pKa = 11.84TSPTRR144 pKa = 11.84IRR146 pKa = 11.84PSAQTPIRR154 pKa = 11.84RR155 pKa = 11.84RR156 pKa = 11.84RR157 pKa = 11.84TTPPTARR164 pKa = 11.84PTTPLTRR171 pKa = 11.84RR172 pKa = 11.84ATVTRR177 pKa = 11.84PRR179 pKa = 11.84PTNPRR184 pKa = 11.84RR185 pKa = 11.84TSPTRR190 pKa = 11.84KK191 pKa = 5.6TTPHH195 pKa = 5.57TTTATSTPRR204 pKa = 11.84TPTTSTPPRR213 pKa = 11.84TARR216 pKa = 11.84PTPTPSPTTIRR227 pKa = 11.84PSTRR231 pKa = 11.84TPIRR235 pKa = 11.84RR236 pKa = 11.84RR237 pKa = 11.84RR238 pKa = 11.84ITLPTARR245 pKa = 11.84PTTLLIRR252 pKa = 11.84RR253 pKa = 11.84TAATSPRR260 pKa = 11.84PTRR263 pKa = 11.84ARR265 pKa = 11.84QISPPRR271 pKa = 11.84TSLTRR276 pKa = 11.84QTTPHH281 pKa = 5.81TTTTPSTPRR290 pKa = 11.84TPTARR295 pKa = 11.84PPRR298 pKa = 11.84RR299 pKa = 11.84TPTRR303 pKa = 11.84SRR305 pKa = 11.84RR306 pKa = 11.84TTPPTTRR313 pKa = 11.84QTTPTIRR320 pKa = 11.84RR321 pKa = 11.84AAATSPRR328 pKa = 11.84PTRR331 pKa = 11.84PRR333 pKa = 11.84QISPPRR339 pKa = 11.84TSLTRR344 pKa = 11.84QTTPHH349 pKa = 5.81TTTTPSTPRR358 pKa = 11.84TPTATPSPTRR368 pKa = 11.84TPTTRR373 pKa = 11.84STPRR377 pKa = 11.84KK378 pKa = 7.72RR379 pKa = 11.84THH381 pKa = 6.88PSTTRR386 pKa = 11.84TRR388 pKa = 11.84TSRR391 pKa = 11.84RR392 pKa = 11.84VPTQQQTSMPATGGPTTLRR411 pKa = 11.84MPGRR415 pKa = 11.84TATRR419 pKa = 11.84KK420 pKa = 9.74RR421 pKa = 11.84QVHH424 pKa = 5.83LPKK427 pKa = 10.24TPTASIPMILLEE439 pKa = 4.31RR440 pKa = 11.84TASPSPLTATPAIHH454 pKa = 7.03RR455 pKa = 11.84AMSTARR461 pKa = 11.84STEE464 pKa = 3.66IVTPTAIRR472 pKa = 11.84ASTKK476 pKa = 9.93ALRR479 pKa = 11.84VTKK482 pKa = 10.11VRR484 pKa = 11.84RR485 pKa = 11.84KK486 pKa = 9.62RR487 pKa = 11.84STRR490 pKa = 11.84ICTITHH496 pKa = 5.04TMARR500 pKa = 3.87
MM1 pKa = 6.74EE2 pKa = 5.03TAPPTHH8 pKa = 6.95PPLLRR13 pKa = 11.84RR14 pKa = 11.84TSPSKK19 pKa = 10.86KK20 pKa = 8.66PWAAPTSSRR29 pKa = 11.84TVEE32 pKa = 4.23GLQQTRR38 pKa = 11.84PPPTRR43 pKa = 11.84TRR45 pKa = 11.84PQTQGRR51 pKa = 11.84LATAPGTQMAPVRR64 pKa = 11.84LIPTLSRR71 pKa = 11.84GTIPASARR79 pKa = 11.84TLTPRR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84TTPPTVRR93 pKa = 11.84PTTPLIRR100 pKa = 11.84RR101 pKa = 11.84ATATRR106 pKa = 11.84PRR108 pKa = 11.84PRR110 pKa = 11.84KK111 pKa = 8.01TSRR114 pKa = 11.84TSPRR118 pKa = 11.84RR119 pKa = 11.84ISPTRR124 pKa = 11.84KK125 pKa = 5.37TTPTASPIRR134 pKa = 11.84TRR136 pKa = 11.84PTRR139 pKa = 11.84TSPTRR144 pKa = 11.84IRR146 pKa = 11.84PSAQTPIRR154 pKa = 11.84RR155 pKa = 11.84RR156 pKa = 11.84RR157 pKa = 11.84TTPPTARR164 pKa = 11.84PTTPLTRR171 pKa = 11.84RR172 pKa = 11.84ATVTRR177 pKa = 11.84PRR179 pKa = 11.84PTNPRR184 pKa = 11.84RR185 pKa = 11.84TSPTRR190 pKa = 11.84KK191 pKa = 5.6TTPHH195 pKa = 5.57TTTATSTPRR204 pKa = 11.84TPTTSTPPRR213 pKa = 11.84TARR216 pKa = 11.84PTPTPSPTTIRR227 pKa = 11.84PSTRR231 pKa = 11.84TPIRR235 pKa = 11.84RR236 pKa = 11.84RR237 pKa = 11.84RR238 pKa = 11.84ITLPTARR245 pKa = 11.84PTTLLIRR252 pKa = 11.84RR253 pKa = 11.84TAATSPRR260 pKa = 11.84PTRR263 pKa = 11.84ARR265 pKa = 11.84QISPPRR271 pKa = 11.84TSLTRR276 pKa = 11.84QTTPHH281 pKa = 5.81TTTTPSTPRR290 pKa = 11.84TPTARR295 pKa = 11.84PPRR298 pKa = 11.84RR299 pKa = 11.84TPTRR303 pKa = 11.84SRR305 pKa = 11.84RR306 pKa = 11.84TTPPTTRR313 pKa = 11.84QTTPTIRR320 pKa = 11.84RR321 pKa = 11.84AAATSPRR328 pKa = 11.84PTRR331 pKa = 11.84PRR333 pKa = 11.84QISPPRR339 pKa = 11.84TSLTRR344 pKa = 11.84QTTPHH349 pKa = 5.81TTTTPSTPRR358 pKa = 11.84TPTATPSPTRR368 pKa = 11.84TPTTRR373 pKa = 11.84STPRR377 pKa = 11.84KK378 pKa = 7.72RR379 pKa = 11.84THH381 pKa = 6.88PSTTRR386 pKa = 11.84TRR388 pKa = 11.84TSRR391 pKa = 11.84RR392 pKa = 11.84VPTQQQTSMPATGGPTTLRR411 pKa = 11.84MPGRR415 pKa = 11.84TATRR419 pKa = 11.84KK420 pKa = 9.74RR421 pKa = 11.84QVHH424 pKa = 5.83LPKK427 pKa = 10.24TPTASIPMILLEE439 pKa = 4.31RR440 pKa = 11.84TASPSPLTATPAIHH454 pKa = 7.03RR455 pKa = 11.84AMSTARR461 pKa = 11.84STEE464 pKa = 3.66IVTPTAIRR472 pKa = 11.84ASTKK476 pKa = 9.93ALRR479 pKa = 11.84VTKK482 pKa = 10.11VRR484 pKa = 11.84RR485 pKa = 11.84KK486 pKa = 9.62RR487 pKa = 11.84STRR490 pKa = 11.84ICTITHH496 pKa = 5.04TMARR500 pKa = 3.87
Molecular weight: 55.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
692431 |
37 |
3083 |
334.5 |
35.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.841 ± 0.077 | 0.819 ± 0.017 |
5.973 ± 0.043 | 5.735 ± 0.053 |
2.879 ± 0.037 | 8.854 ± 0.047 |
2.008 ± 0.023 | 4.653 ± 0.04 |
2.393 ± 0.035 | 9.278 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.201 ± 0.024 | 2.248 ± 0.033 |
5.33 ± 0.048 | 2.912 ± 0.03 |
7.147 ± 0.064 | 6.629 ± 0.049 |
6.023 ± 0.05 | 8.36 ± 0.053 |
1.517 ± 0.026 | 2.202 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
