
Marinobacterium sp. AK27
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Oceanospirillaceae; Marinobacterium; unclassified Marinobacterium
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
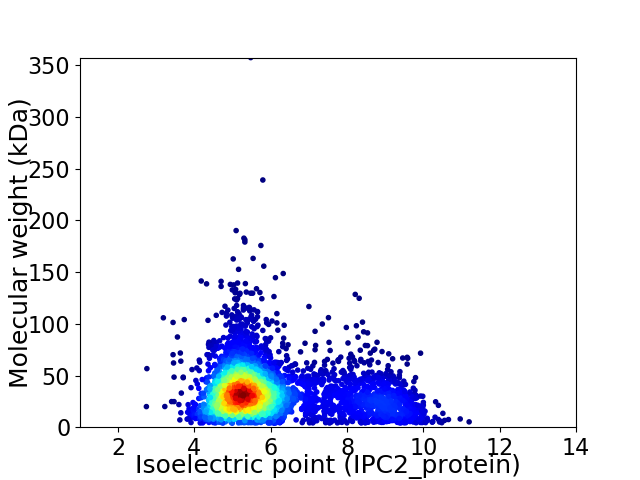
Virtual 2D-PAGE plot for 4141 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A081G436|A0A081G436_9GAMM Biotin carboxylase OS=Marinobacterium sp. AK27 OX=1232683 GN=ADIMK_0498 PE=4 SV=1
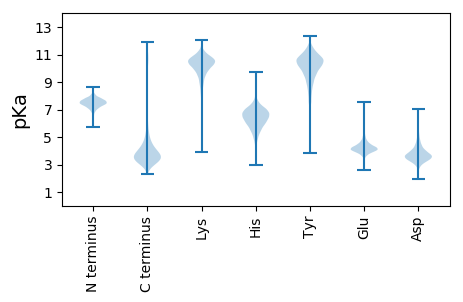
MM1 pKa = 7.35NRR3 pKa = 11.84QLQGGMRR10 pKa = 11.84KK11 pKa = 7.82QQQGVVLVLVAIAMLALLAMAGLALDD37 pKa = 4.62GGHH40 pKa = 7.46LLLNKK45 pKa = 9.09TRR47 pKa = 11.84LQNAVDD53 pKa = 3.72AAALSAARR61 pKa = 11.84TIADD65 pKa = 3.92FPKK68 pKa = 9.05DD69 pKa = 3.46TAVGTVHH76 pKa = 6.56TAAEE80 pKa = 4.24ASALAAFLSNLGLDD94 pKa = 5.01DD95 pKa = 4.46SAEE98 pKa = 4.08LNDD101 pKa = 4.65AYY103 pKa = 10.6ASSGSPLQVQFSTTLNPFVDD123 pKa = 3.87SATALPYY130 pKa = 10.62VRR132 pKa = 11.84VIAQDD137 pKa = 3.5LQLDD141 pKa = 3.77VWFLQVVGMADD152 pKa = 3.45KK153 pKa = 10.24PVSASAVAGPIGLNANTCDD172 pKa = 3.52ILPILACGCDD182 pKa = 3.26TDD184 pKa = 4.72GTYY187 pKa = 10.26TPDD190 pKa = 5.01NPDD193 pKa = 4.38DD194 pKa = 4.55DD195 pKa = 6.14CSDD198 pKa = 3.43SNTYY202 pKa = 10.23FGYY205 pKa = 9.95PDD207 pKa = 3.75NDD209 pKa = 3.69NNDD212 pKa = 3.43VEE214 pKa = 4.65SLGDD218 pKa = 3.27ISVIKK223 pKa = 10.3LAGGSGSDD231 pKa = 3.25VGPGNFRR238 pKa = 11.84LLRR241 pKa = 11.84LDD243 pKa = 3.36GTGGDD248 pKa = 4.32DD249 pKa = 5.55LRR251 pKa = 11.84DD252 pKa = 3.56ALAGVGDD259 pKa = 3.87SCANFGDD266 pKa = 4.71DD267 pKa = 4.1SAADD271 pKa = 3.74TEE273 pKa = 4.73PGNKK277 pKa = 9.43SGPVGQGLNTRR288 pKa = 11.84LGIYY292 pKa = 9.2QGPVDD297 pKa = 4.65PTTAPADD304 pKa = 3.74EE305 pKa = 4.34VTYY308 pKa = 9.48TYY310 pKa = 11.37YY311 pKa = 10.83SGSTPAGPTPVGPNPLVLDD330 pKa = 4.17GLDD333 pKa = 3.65DD334 pKa = 3.7QGVPVIKK341 pKa = 10.34RR342 pKa = 11.84QDD344 pKa = 3.15GSTSIDD350 pKa = 3.29GLFDD354 pKa = 3.46YY355 pKa = 10.92QDD357 pKa = 3.65YY358 pKa = 10.99LDD360 pKa = 5.33EE361 pKa = 4.39YY362 pKa = 9.11TSNYY366 pKa = 9.18PDD368 pKa = 3.86CQGSGCRR375 pKa = 11.84RR376 pKa = 11.84RR377 pKa = 11.84EE378 pKa = 3.82LVLPIGDD385 pKa = 4.01CTGSISGTSEE395 pKa = 3.78DD396 pKa = 3.45VHH398 pKa = 8.0VYY400 pKa = 10.47SVGCFFLMQNVDD412 pKa = 3.27VGTDD416 pKa = 3.31AEE418 pKa = 4.59VFGQFFTEE426 pKa = 5.08SEE428 pKa = 4.19GCDD431 pKa = 3.32AIGTFTSTPTSDD443 pKa = 3.47PVPIKK448 pKa = 10.27IVLYY452 pKa = 10.16RR453 pKa = 11.84DD454 pKa = 3.47SDD456 pKa = 3.87SGDD459 pKa = 3.29SS460 pKa = 3.44
MM1 pKa = 7.35NRR3 pKa = 11.84QLQGGMRR10 pKa = 11.84KK11 pKa = 7.82QQQGVVLVLVAIAMLALLAMAGLALDD37 pKa = 4.62GGHH40 pKa = 7.46LLLNKK45 pKa = 9.09TRR47 pKa = 11.84LQNAVDD53 pKa = 3.72AAALSAARR61 pKa = 11.84TIADD65 pKa = 3.92FPKK68 pKa = 9.05DD69 pKa = 3.46TAVGTVHH76 pKa = 6.56TAAEE80 pKa = 4.24ASALAAFLSNLGLDD94 pKa = 5.01DD95 pKa = 4.46SAEE98 pKa = 4.08LNDD101 pKa = 4.65AYY103 pKa = 10.6ASSGSPLQVQFSTTLNPFVDD123 pKa = 3.87SATALPYY130 pKa = 10.62VRR132 pKa = 11.84VIAQDD137 pKa = 3.5LQLDD141 pKa = 3.77VWFLQVVGMADD152 pKa = 3.45KK153 pKa = 10.24PVSASAVAGPIGLNANTCDD172 pKa = 3.52ILPILACGCDD182 pKa = 3.26TDD184 pKa = 4.72GTYY187 pKa = 10.26TPDD190 pKa = 5.01NPDD193 pKa = 4.38DD194 pKa = 4.55DD195 pKa = 6.14CSDD198 pKa = 3.43SNTYY202 pKa = 10.23FGYY205 pKa = 9.95PDD207 pKa = 3.75NDD209 pKa = 3.69NNDD212 pKa = 3.43VEE214 pKa = 4.65SLGDD218 pKa = 3.27ISVIKK223 pKa = 10.3LAGGSGSDD231 pKa = 3.25VGPGNFRR238 pKa = 11.84LLRR241 pKa = 11.84LDD243 pKa = 3.36GTGGDD248 pKa = 4.32DD249 pKa = 5.55LRR251 pKa = 11.84DD252 pKa = 3.56ALAGVGDD259 pKa = 3.87SCANFGDD266 pKa = 4.71DD267 pKa = 4.1SAADD271 pKa = 3.74TEE273 pKa = 4.73PGNKK277 pKa = 9.43SGPVGQGLNTRR288 pKa = 11.84LGIYY292 pKa = 9.2QGPVDD297 pKa = 4.65PTTAPADD304 pKa = 3.74EE305 pKa = 4.34VTYY308 pKa = 9.48TYY310 pKa = 11.37YY311 pKa = 10.83SGSTPAGPTPVGPNPLVLDD330 pKa = 4.17GLDD333 pKa = 3.65DD334 pKa = 3.7QGVPVIKK341 pKa = 10.34RR342 pKa = 11.84QDD344 pKa = 3.15GSTSIDD350 pKa = 3.29GLFDD354 pKa = 3.46YY355 pKa = 10.92QDD357 pKa = 3.65YY358 pKa = 10.99LDD360 pKa = 5.33EE361 pKa = 4.39YY362 pKa = 9.11TSNYY366 pKa = 9.18PDD368 pKa = 3.86CQGSGCRR375 pKa = 11.84RR376 pKa = 11.84RR377 pKa = 11.84EE378 pKa = 3.82LVLPIGDD385 pKa = 4.01CTGSISGTSEE395 pKa = 3.78DD396 pKa = 3.45VHH398 pKa = 8.0VYY400 pKa = 10.47SVGCFFLMQNVDD412 pKa = 3.27VGTDD416 pKa = 3.31AEE418 pKa = 4.59VFGQFFTEE426 pKa = 5.08SEE428 pKa = 4.19GCDD431 pKa = 3.32AIGTFTSTPTSDD443 pKa = 3.47PVPIKK448 pKa = 10.27IVLYY452 pKa = 10.16RR453 pKa = 11.84DD454 pKa = 3.47SDD456 pKa = 3.87SGDD459 pKa = 3.29SS460 pKa = 3.44
Molecular weight: 47.85 kDa
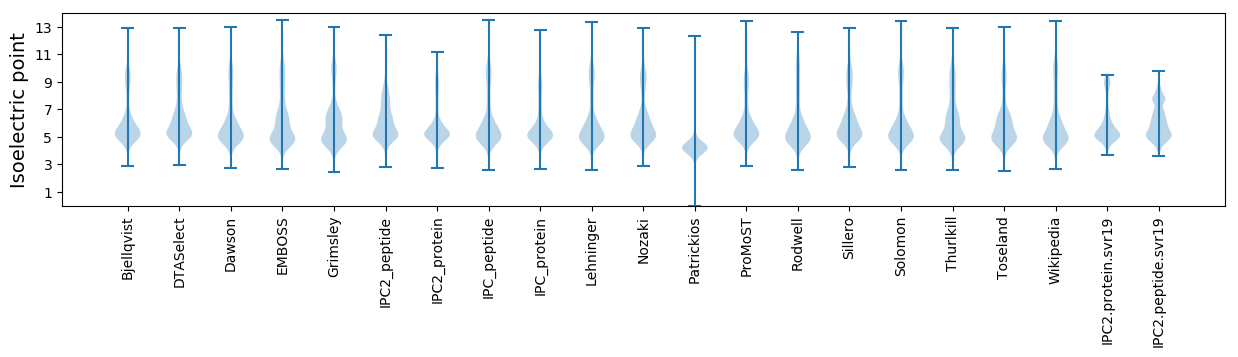
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A081FVI1|A0A081FVI1_9GAMM ATP synthase subunit alpha OS=Marinobacterium sp. AK27 OX=1232683 GN=atpA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.91GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.45NGRR28 pKa = 11.84QVLNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.89SLTVV44 pKa = 3.12
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.91GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.45NGRR28 pKa = 11.84QVLNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.89SLTVV44 pKa = 3.12
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1346198 |
37 |
3254 |
325.1 |
35.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.954 ± 0.043 | 1.077 ± 0.014 |
5.638 ± 0.03 | 6.459 ± 0.034 |
3.649 ± 0.023 | 7.662 ± 0.034 |
2.248 ± 0.021 | 5.557 ± 0.028 |
3.583 ± 0.025 | 11.341 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.613 ± 0.017 | 3.126 ± 0.019 |
4.527 ± 0.024 | 4.217 ± 0.027 |
6.303 ± 0.037 | 6.136 ± 0.027 |
4.923 ± 0.026 | 7.012 ± 0.031 |
1.294 ± 0.014 | 2.681 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
