
Sheep faeces associated smacovirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus; Porprismacovirus sheas3
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
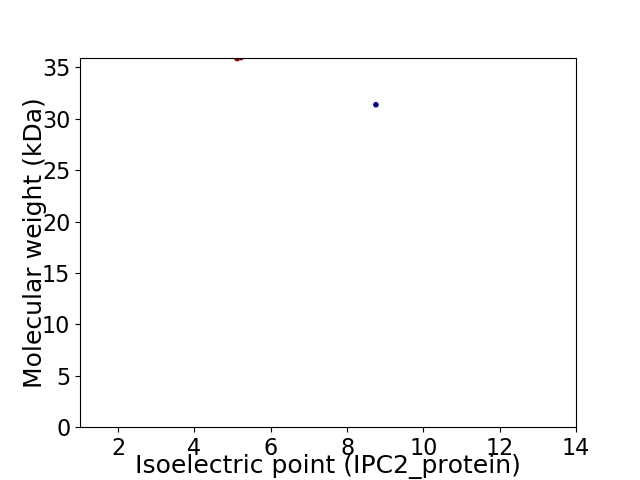
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A168MFI8|A0A168MFI8_9VIRU Replication associated protein OS=Sheep faeces associated smacovirus 3 OX=1843758 PE=4 SV=1
MM1 pKa = 7.16VFVKK5 pKa = 10.43VRR7 pKa = 11.84EE8 pKa = 4.3TYY10 pKa = 10.49DD11 pKa = 3.1LHH13 pKa = 5.18TLKK16 pKa = 11.12GKK18 pKa = 9.6MSVIGIHH25 pKa = 5.87TPSDD29 pKa = 3.73SIIARR34 pKa = 11.84NYY36 pKa = 9.5PGLLMQCKK44 pKa = 9.96AYY46 pKa = 10.43RR47 pKa = 11.84PVKK50 pKa = 10.41ADD52 pKa = 3.1VRR54 pKa = 11.84LACASVLPLDD64 pKa = 4.08PQGVGTTADD73 pKa = 3.49DD74 pKa = 3.77VAPEE78 pKa = 4.34DD79 pKa = 3.68VFNPILYY86 pKa = 9.58KK87 pKa = 10.71ALSNFGMSQIDD98 pKa = 3.84AYY100 pKa = 11.06VNSAGYY106 pKa = 10.09DD107 pKa = 3.38IIGNTLDD114 pKa = 4.02ASNTGIDD121 pKa = 3.44ATDD124 pKa = 4.62DD125 pKa = 3.34FDD127 pKa = 5.92LYY129 pKa = 11.44YY130 pKa = 11.28GLLSQTHH137 pKa = 5.37EE138 pKa = 4.23WKK140 pKa = 10.26HH141 pKa = 6.12ANPQQGLMMNDD152 pKa = 4.21LVPLVYY158 pKa = 9.02EE159 pKa = 5.03TYY161 pKa = 10.85QSIGDD166 pKa = 3.9NGRR169 pKa = 11.84GGADD173 pKa = 3.45NPHH176 pKa = 6.03VAIKK180 pKa = 10.35PDD182 pKa = 3.49DD183 pKa = 4.53TIPTGQIGNISVQTFRR199 pKa = 11.84GKK201 pKa = 10.06AHH203 pKa = 7.72PIPFLNCTVPVANSGNVDD221 pKa = 2.94TRR223 pKa = 11.84QNGFIGSGYY232 pKa = 9.75PNAQVGIPAPKK243 pKa = 9.31IYY245 pKa = 10.23CAAILVPPSRR255 pKa = 11.84LHH257 pKa = 5.42QLFFRR262 pKa = 11.84MVCEE266 pKa = 3.97WTLEE270 pKa = 3.86FSMIRR275 pKa = 11.84PLGEE279 pKa = 3.63ITSWAGLKK287 pKa = 10.46QMGSTSHH294 pKa = 6.46FMSYY298 pKa = 10.59DD299 pKa = 3.36YY300 pKa = 11.16SAEE303 pKa = 4.16SKK305 pKa = 10.02DD306 pKa = 3.71TVFKK310 pKa = 11.06DD311 pKa = 3.29VTDD314 pKa = 4.23LVDD317 pKa = 3.25VTQDD321 pKa = 3.15SGIKK325 pKa = 10.09KK326 pKa = 10.23VMM328 pKa = 3.67
MM1 pKa = 7.16VFVKK5 pKa = 10.43VRR7 pKa = 11.84EE8 pKa = 4.3TYY10 pKa = 10.49DD11 pKa = 3.1LHH13 pKa = 5.18TLKK16 pKa = 11.12GKK18 pKa = 9.6MSVIGIHH25 pKa = 5.87TPSDD29 pKa = 3.73SIIARR34 pKa = 11.84NYY36 pKa = 9.5PGLLMQCKK44 pKa = 9.96AYY46 pKa = 10.43RR47 pKa = 11.84PVKK50 pKa = 10.41ADD52 pKa = 3.1VRR54 pKa = 11.84LACASVLPLDD64 pKa = 4.08PQGVGTTADD73 pKa = 3.49DD74 pKa = 3.77VAPEE78 pKa = 4.34DD79 pKa = 3.68VFNPILYY86 pKa = 9.58KK87 pKa = 10.71ALSNFGMSQIDD98 pKa = 3.84AYY100 pKa = 11.06VNSAGYY106 pKa = 10.09DD107 pKa = 3.38IIGNTLDD114 pKa = 4.02ASNTGIDD121 pKa = 3.44ATDD124 pKa = 4.62DD125 pKa = 3.34FDD127 pKa = 5.92LYY129 pKa = 11.44YY130 pKa = 11.28GLLSQTHH137 pKa = 5.37EE138 pKa = 4.23WKK140 pKa = 10.26HH141 pKa = 6.12ANPQQGLMMNDD152 pKa = 4.21LVPLVYY158 pKa = 9.02EE159 pKa = 5.03TYY161 pKa = 10.85QSIGDD166 pKa = 3.9NGRR169 pKa = 11.84GGADD173 pKa = 3.45NPHH176 pKa = 6.03VAIKK180 pKa = 10.35PDD182 pKa = 3.49DD183 pKa = 4.53TIPTGQIGNISVQTFRR199 pKa = 11.84GKK201 pKa = 10.06AHH203 pKa = 7.72PIPFLNCTVPVANSGNVDD221 pKa = 2.94TRR223 pKa = 11.84QNGFIGSGYY232 pKa = 9.75PNAQVGIPAPKK243 pKa = 9.31IYY245 pKa = 10.23CAAILVPPSRR255 pKa = 11.84LHH257 pKa = 5.42QLFFRR262 pKa = 11.84MVCEE266 pKa = 3.97WTLEE270 pKa = 3.86FSMIRR275 pKa = 11.84PLGEE279 pKa = 3.63ITSWAGLKK287 pKa = 10.46QMGSTSHH294 pKa = 6.46FMSYY298 pKa = 10.59DD299 pKa = 3.36YY300 pKa = 11.16SAEE303 pKa = 4.16SKK305 pKa = 10.02DD306 pKa = 3.71TVFKK310 pKa = 11.06DD311 pKa = 3.29VTDD314 pKa = 4.23LVDD317 pKa = 3.25VTQDD321 pKa = 3.15SGIKK325 pKa = 10.09KK326 pKa = 10.23VMM328 pKa = 3.67
Molecular weight: 35.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A168MFI8|A0A168MFI8_9VIRU Replication associated protein OS=Sheep faeces associated smacovirus 3 OX=1843758 PE=4 SV=1
MM1 pKa = 7.31LTMPWTGQSKK11 pKa = 10.5RR12 pKa = 11.84MINLYY17 pKa = 8.17ITRR20 pKa = 11.84NDD22 pKa = 2.59IHH24 pKa = 6.41EE25 pKa = 4.53WIIGYY30 pKa = 8.82EE31 pKa = 4.09VGRR34 pKa = 11.84DD35 pKa = 3.96GYY37 pKa = 10.66RR38 pKa = 11.84HH39 pKa = 4.43IHH41 pKa = 4.77VRR43 pKa = 11.84FNGRR47 pKa = 11.84GDD49 pKa = 3.44FGDD52 pKa = 3.57VQRR55 pKa = 11.84AFPGAHH61 pKa = 6.06IEE63 pKa = 4.18EE64 pKa = 4.85GTTMEE69 pKa = 4.18TEE71 pKa = 4.11YY72 pKa = 10.79EE73 pKa = 4.2KK74 pKa = 11.12KK75 pKa = 10.24DD76 pKa = 3.43GHH78 pKa = 5.69FVSYY82 pKa = 10.57EE83 pKa = 3.82DD84 pKa = 3.66SPDD87 pKa = 3.37VLRR90 pKa = 11.84CRR92 pKa = 11.84FGKK95 pKa = 10.44LRR97 pKa = 11.84DD98 pKa = 3.47HH99 pKa = 6.13QRR101 pKa = 11.84RR102 pKa = 11.84VVKK105 pKa = 10.47LLEE108 pKa = 4.33KK109 pKa = 10.65QSDD112 pKa = 3.34RR113 pKa = 11.84GILCWYY119 pKa = 9.96DD120 pKa = 3.22EE121 pKa = 4.35TGSIGKK127 pKa = 9.51SFTCRR132 pKa = 11.84HH133 pKa = 4.44LVEE136 pKa = 4.39RR137 pKa = 11.84RR138 pKa = 11.84RR139 pKa = 11.84AYY141 pKa = 9.74YY142 pKa = 10.16VPPTVDD148 pKa = 2.86NVKK151 pKa = 10.64QIIQWVCSGYY161 pKa = 9.86QRR163 pKa = 11.84QRR165 pKa = 11.84YY166 pKa = 8.83LIIDD170 pKa = 4.1IPRR173 pKa = 11.84SAEE176 pKa = 3.97WNKK179 pKa = 10.73SLYY182 pKa = 9.85TGLEE186 pKa = 4.07AIKK189 pKa = 10.75DD190 pKa = 3.53GLIYY194 pKa = 9.15DD195 pKa = 3.92TRR197 pKa = 11.84YY198 pKa = 9.47SAKK201 pKa = 10.33LRR203 pKa = 11.84DD204 pKa = 3.18IWGVKK209 pKa = 9.72ILVLTNSLPNLDD221 pKa = 5.19ALSQDD226 pKa = 2.08RR227 pKa = 11.84WMIINRR233 pKa = 11.84DD234 pKa = 3.59GKK236 pKa = 10.45DD237 pKa = 3.12VTEE240 pKa = 4.16WARR243 pKa = 11.84IEE245 pKa = 4.31SKK247 pKa = 10.66KK248 pKa = 9.79RR249 pKa = 11.84AKK251 pKa = 10.26KK252 pKa = 10.37KK253 pKa = 10.27ADD255 pKa = 3.23EE256 pKa = 4.53GGKK259 pKa = 10.23KK260 pKa = 8.26KK261 pKa = 9.0TKK263 pKa = 10.11KK264 pKa = 10.35SEE266 pKa = 3.89EE267 pKa = 3.94
MM1 pKa = 7.31LTMPWTGQSKK11 pKa = 10.5RR12 pKa = 11.84MINLYY17 pKa = 8.17ITRR20 pKa = 11.84NDD22 pKa = 2.59IHH24 pKa = 6.41EE25 pKa = 4.53WIIGYY30 pKa = 8.82EE31 pKa = 4.09VGRR34 pKa = 11.84DD35 pKa = 3.96GYY37 pKa = 10.66RR38 pKa = 11.84HH39 pKa = 4.43IHH41 pKa = 4.77VRR43 pKa = 11.84FNGRR47 pKa = 11.84GDD49 pKa = 3.44FGDD52 pKa = 3.57VQRR55 pKa = 11.84AFPGAHH61 pKa = 6.06IEE63 pKa = 4.18EE64 pKa = 4.85GTTMEE69 pKa = 4.18TEE71 pKa = 4.11YY72 pKa = 10.79EE73 pKa = 4.2KK74 pKa = 11.12KK75 pKa = 10.24DD76 pKa = 3.43GHH78 pKa = 5.69FVSYY82 pKa = 10.57EE83 pKa = 3.82DD84 pKa = 3.66SPDD87 pKa = 3.37VLRR90 pKa = 11.84CRR92 pKa = 11.84FGKK95 pKa = 10.44LRR97 pKa = 11.84DD98 pKa = 3.47HH99 pKa = 6.13QRR101 pKa = 11.84RR102 pKa = 11.84VVKK105 pKa = 10.47LLEE108 pKa = 4.33KK109 pKa = 10.65QSDD112 pKa = 3.34RR113 pKa = 11.84GILCWYY119 pKa = 9.96DD120 pKa = 3.22EE121 pKa = 4.35TGSIGKK127 pKa = 9.51SFTCRR132 pKa = 11.84HH133 pKa = 4.44LVEE136 pKa = 4.39RR137 pKa = 11.84RR138 pKa = 11.84RR139 pKa = 11.84AYY141 pKa = 9.74YY142 pKa = 10.16VPPTVDD148 pKa = 2.86NVKK151 pKa = 10.64QIIQWVCSGYY161 pKa = 9.86QRR163 pKa = 11.84QRR165 pKa = 11.84YY166 pKa = 8.83LIIDD170 pKa = 4.1IPRR173 pKa = 11.84SAEE176 pKa = 3.97WNKK179 pKa = 10.73SLYY182 pKa = 9.85TGLEE186 pKa = 4.07AIKK189 pKa = 10.75DD190 pKa = 3.53GLIYY194 pKa = 9.15DD195 pKa = 3.92TRR197 pKa = 11.84YY198 pKa = 9.47SAKK201 pKa = 10.33LRR203 pKa = 11.84DD204 pKa = 3.18IWGVKK209 pKa = 9.72ILVLTNSLPNLDD221 pKa = 5.19ALSQDD226 pKa = 2.08RR227 pKa = 11.84WMIINRR233 pKa = 11.84DD234 pKa = 3.59GKK236 pKa = 10.45DD237 pKa = 3.12VTEE240 pKa = 4.16WARR243 pKa = 11.84IEE245 pKa = 4.31SKK247 pKa = 10.66KK248 pKa = 9.79RR249 pKa = 11.84AKK251 pKa = 10.26KK252 pKa = 10.37KK253 pKa = 10.27ADD255 pKa = 3.23EE256 pKa = 4.53GGKK259 pKa = 10.23KK260 pKa = 8.26KK261 pKa = 9.0TKK263 pKa = 10.11KK264 pKa = 10.35SEE266 pKa = 3.89EE267 pKa = 3.94
Molecular weight: 31.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
595 |
267 |
328 |
297.5 |
33.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.546 ± 1.161 | 1.513 ± 0.009 |
7.731 ± 0.155 | 4.37 ± 1.529 |
3.025 ± 0.502 | 8.067 ± 0.13 |
2.521 ± 0.065 | 7.227 ± 0.411 |
6.387 ± 1.436 | 7.059 ± 0.205 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.689 ± 0.526 | 4.034 ± 0.669 |
4.706 ± 1.343 | 3.866 ± 0.319 |
6.05 ± 2.377 | 6.05 ± 0.52 |
5.882 ± 0.412 | 6.891 ± 0.82 |
1.849 ± 0.74 | 4.538 ± 0.213 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
