
Acidipila dinghuensis
Taxonomy: cellular organisms; Bacteria; Acidobacteria; Acidobacteriia; Acidobacteriales; Acidobacteriaceae; Acidipila
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
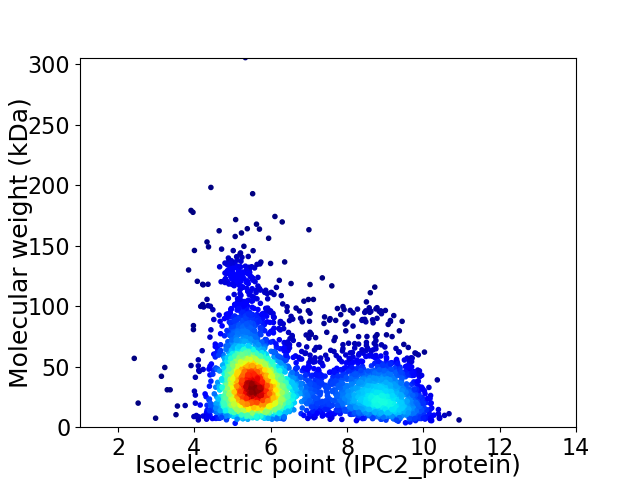
Virtual 2D-PAGE plot for 3909 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q1SDS9|A0A4Q1SDS9_9BACT NAD(P)-dependent alcohol dehydrogenase OS=Acidipila dinghuensis OX=1560006 GN=ESZ00_11500 PE=3 SV=1
MM1 pKa = 7.6ASSASIVSLLEE12 pKa = 3.96SLGVPILQEE21 pKa = 3.87VADD24 pKa = 4.05AALGQGVLTLSDD36 pKa = 4.01DD37 pKa = 3.87ASGLTVGVTLSSGQSLHH54 pKa = 6.98ADD56 pKa = 3.31DD57 pKa = 4.19VTFMGSQGFSGHH69 pKa = 6.42FYY71 pKa = 11.29VDD73 pKa = 3.55GLDD76 pKa = 4.06ANPLSATLAGGFAIALTAFDD96 pKa = 3.56VTIANGGMAASTIGGALTIPFFTDD120 pKa = 2.88EE121 pKa = 4.31NGNPEE126 pKa = 4.15TLDD129 pKa = 3.44VEE131 pKa = 5.0VSTKK135 pKa = 10.15QDD137 pKa = 2.76GSLCITLAATTSSPTTPDD155 pKa = 2.92GLVQLVYY162 pKa = 10.8NLPGGLGSVEE172 pKa = 3.83IDD174 pKa = 3.43VASIEE179 pKa = 4.18FDD181 pKa = 3.17EE182 pKa = 5.64SAGTWTIIISGNLIITVPGLNWPSIEE208 pKa = 4.44LRR210 pKa = 11.84GLGIDD215 pKa = 3.45SKK217 pKa = 11.9GNISLQGGWINLPNQMALDD236 pKa = 4.39FYY238 pKa = 10.88GFSIALQALGFGSDD252 pKa = 5.24ASGKK256 pKa = 9.18WIGFNGDD263 pKa = 2.92INLVEE268 pKa = 4.34GLSLGGSVQGLKK280 pKa = 10.9VNLTTGSISFNGVGISFEE298 pKa = 4.07IPGVITIDD306 pKa = 3.57GEE308 pKa = 3.88IDD310 pKa = 4.07HH311 pKa = 6.76IHH313 pKa = 6.35VDD315 pKa = 3.6ANTPNDD321 pKa = 3.87LVPYY325 pKa = 10.1GLMPSIFNSIAPVGPTGPKK344 pKa = 10.34SIDD347 pKa = 3.3LFAGSVKK354 pKa = 10.54VQVLPDD360 pKa = 3.64EE361 pKa = 4.71MDD363 pKa = 3.95LEE365 pKa = 4.28VDD367 pKa = 3.41ANFIVGNFGGQSVFFLALDD386 pKa = 3.49VEE388 pKa = 4.84LPVGIPIFMDD398 pKa = 3.51VSLYY402 pKa = 10.87GLSGLVATGLDD413 pKa = 4.23PDD415 pKa = 4.87PEE417 pKa = 5.62PNDD420 pKa = 4.23TWWQWYY426 pKa = 9.19KK427 pKa = 11.2YY428 pKa = 9.17PAGTSGPQLNATPDD442 pKa = 3.42YY443 pKa = 10.88SATDD447 pKa = 3.67FTKK450 pKa = 10.28WLVPQQGAFAIGGGCTIGTSADD472 pKa = 3.36DD473 pKa = 4.91GYY475 pKa = 9.38TVSAAIMLVILMPGPVISLIGKK497 pKa = 9.87ANILSKK503 pKa = 10.47RR504 pKa = 11.84ISGADD509 pKa = 2.97QDD511 pKa = 4.92ANFEE515 pKa = 4.29AMATYY520 pKa = 10.38DD521 pKa = 3.98GNSGTFDD528 pKa = 3.45LTIDD532 pKa = 3.58AQYY535 pKa = 10.59SIPVVLDD542 pKa = 3.43IEE544 pKa = 4.56ATAEE548 pKa = 4.21LYY550 pKa = 10.96VDD552 pKa = 4.06ATAGEE557 pKa = 4.48WFFAMGKK564 pKa = 9.0PPHH567 pKa = 5.33TQRR570 pKa = 11.84VSARR574 pKa = 11.84IFDD577 pKa = 3.72IFEE580 pKa = 3.78TDD582 pKa = 2.71AYY584 pKa = 10.55FVVSNAGLITGTWTGYY600 pKa = 8.9QGSWSFGPLSASLDD614 pKa = 3.57AYY616 pKa = 10.85LATLAAIQWSPLQIAGGIEE635 pKa = 3.93LHH637 pKa = 7.09GSVQLSAFGIGVGISADD654 pKa = 3.59ALLEE658 pKa = 4.38GCAPNPFWVHH668 pKa = 5.99GEE670 pKa = 3.94LSVEE674 pKa = 5.31LDD676 pKa = 4.43LPWPLPNVGGTISLSWGGDD695 pKa = 3.47DD696 pKa = 5.37GSIPPVPLALSHH708 pKa = 7.03IDD710 pKa = 3.75AALSDD715 pKa = 4.33HH716 pKa = 6.84GDD718 pKa = 3.73SAGKK722 pKa = 8.64PASDD726 pKa = 3.55HH727 pKa = 5.97YY728 pKa = 11.69VLLAHH733 pKa = 7.43RR734 pKa = 11.84SNGPFPDD741 pKa = 3.62LTVQYY746 pKa = 10.21DD747 pKa = 3.49NPQAPGILDD756 pKa = 3.54LTGTALTNWQNRR768 pKa = 11.84VSSGNLADD776 pKa = 4.8MLPDD780 pKa = 3.45MTPDD784 pKa = 3.18NTSRR788 pKa = 11.84TQMAPVLPQDD798 pKa = 3.26THH800 pKa = 6.18FVLNFSHH807 pKa = 6.96PVVDD811 pKa = 3.89MSGGFNNALPQAQLPPEE828 pKa = 4.6VVTTPPPPPPTEE840 pKa = 3.87VGKK843 pKa = 10.61DD844 pKa = 3.29DD845 pKa = 4.09MSNINPNPPSVQFLIRR861 pKa = 11.84HH862 pKa = 5.09SLIDD866 pKa = 3.36VSLYY870 pKa = 10.42EE871 pKa = 4.18WVGGAWSLVASIPPVPPTEE890 pKa = 4.73ASAQADD896 pKa = 3.78TTYY899 pKa = 11.45LSGVWLAPTSATQIQLKK916 pKa = 8.58VVPWMMVEE924 pKa = 4.27GEE926 pKa = 4.31FWTVSWSSTSTPQNYY941 pKa = 7.4GTSFNNQGLDD951 pKa = 3.59FQTTNTQPAAIGTGNAPGLQQGLAFTNAGGSQQPSVAITFPQPEE995 pKa = 4.44VLSSLTAVVAVDD1007 pKa = 3.68EE1008 pKa = 5.04GEE1010 pKa = 4.42LLYY1013 pKa = 10.75AYY1015 pKa = 8.01NAPTCVGDD1023 pKa = 4.4GVTLTPTAQSWDD1035 pKa = 3.88SNFMQWTLTFDD1046 pKa = 3.91PEE1048 pKa = 4.06ATPISTLTLSMQNTSGATLVLYY1070 pKa = 10.7ALDD1073 pKa = 3.71YY1074 pKa = 8.63TVPPTAMAILPKK1086 pKa = 10.67APATYY1091 pKa = 10.87ALRR1094 pKa = 11.84VTTKK1098 pKa = 10.26IEE1100 pKa = 3.73AARR1103 pKa = 11.84VNGGTPQYY1111 pKa = 9.35QTAPDD1116 pKa = 4.21GSPIVEE1122 pKa = 4.22FAYY1125 pKa = 10.18FQTACGPGIAEE1136 pKa = 4.61IGASIPNTSGLPSTPTNTTPVPAEE1160 pKa = 3.76AANFPQLAVNCSTMQQPFTSFPLGGAIDD1188 pKa = 5.15DD1189 pKa = 4.16LTTYY1193 pKa = 7.25TQWSWPLNGAVTAYY1207 pKa = 10.33YY1208 pKa = 10.29GYY1210 pKa = 10.68DD1211 pKa = 3.41VNVEE1215 pKa = 4.09FCEE1218 pKa = 4.66SYY1220 pKa = 11.62VNALYY1225 pKa = 10.57VAAGGRR1231 pKa = 11.84GIANSLHH1238 pKa = 5.59FRR1240 pKa = 11.84CVDD1243 pKa = 3.27RR1244 pKa = 11.84NQNHH1248 pKa = 7.23LLLQTLAIHH1257 pKa = 6.1VPSIPQQSALVATALTVPLPSVIQPSDD1284 pKa = 3.37QDD1286 pKa = 3.61GLLTLTGAQLAGLEE1300 pKa = 4.0KK1301 pKa = 10.33RR1302 pKa = 11.84ALVKK1306 pKa = 9.99IDD1308 pKa = 4.82SNLQPDD1314 pKa = 4.39TAASLQNTAITNVSQLTGPGIQQLNPGIIGIILKK1348 pKa = 9.69EE1349 pKa = 4.03RR1350 pKa = 11.84QEE1352 pKa = 3.97QEE1354 pKa = 3.39AAATARR1360 pKa = 11.84QLWFQPLAPQMQYY1373 pKa = 9.79TLDD1376 pKa = 3.63VVAGSWLGATSDD1388 pKa = 4.36FYY1390 pKa = 10.93PAAGAGTGSLEE1401 pKa = 4.48EE1402 pKa = 4.47IFSASDD1408 pKa = 3.96AISLLKK1414 pKa = 10.67DD1415 pKa = 3.29LEE1417 pKa = 4.25NWYY1420 pKa = 8.14TAQAALTTLEE1430 pKa = 4.18RR1431 pKa = 11.84VTFTTSRR1438 pKa = 11.84YY1439 pKa = 8.54ATFSSQMSNAADD1451 pKa = 3.49QLAGVAGTAPLRR1463 pKa = 11.84HH1464 pKa = 6.33YY1465 pKa = 7.76NTSVDD1470 pKa = 3.77VNTWLADD1477 pKa = 3.48AANGYY1482 pKa = 9.14SAFTTAQTIYY1492 pKa = 8.17TQQRR1496 pKa = 11.84ATLAGVVAQFDD1507 pKa = 3.84PRR1509 pKa = 11.84ADD1511 pKa = 3.77DD1512 pKa = 4.39LAPGAPWPDD1521 pKa = 3.57NGNHH1525 pKa = 6.49ALVTARR1531 pKa = 11.84EE1532 pKa = 4.32ATSQAWQALQQTSVPVFDD1550 pKa = 5.05ALIGALGRR1558 pKa = 11.84ADD1560 pKa = 3.44LTSTQKK1566 pKa = 10.51PVAVPDD1572 pKa = 3.7TEE1574 pKa = 4.21MTLFLDD1580 pKa = 3.49PTGFDD1585 pKa = 2.87IRR1587 pKa = 11.84AILLEE1592 pKa = 4.41SPEE1595 pKa = 4.03ALPWQRR1601 pKa = 11.84IWQWIQLTPISPSSLGLKK1619 pKa = 10.12QIDD1622 pKa = 3.78VLWNSDD1628 pKa = 3.21NTRR1631 pKa = 11.84GLILPLGSPRR1641 pKa = 11.84GRR1643 pKa = 11.84YY1644 pKa = 6.25TLSITFQGNLGAEE1657 pKa = 4.43APCITLNGLPVTEE1670 pKa = 4.44AATIGVLSLGAVIHH1684 pKa = 6.43KK1685 pKa = 9.38PRR1687 pKa = 11.84NLL1689 pKa = 3.35
MM1 pKa = 7.6ASSASIVSLLEE12 pKa = 3.96SLGVPILQEE21 pKa = 3.87VADD24 pKa = 4.05AALGQGVLTLSDD36 pKa = 4.01DD37 pKa = 3.87ASGLTVGVTLSSGQSLHH54 pKa = 6.98ADD56 pKa = 3.31DD57 pKa = 4.19VTFMGSQGFSGHH69 pKa = 6.42FYY71 pKa = 11.29VDD73 pKa = 3.55GLDD76 pKa = 4.06ANPLSATLAGGFAIALTAFDD96 pKa = 3.56VTIANGGMAASTIGGALTIPFFTDD120 pKa = 2.88EE121 pKa = 4.31NGNPEE126 pKa = 4.15TLDD129 pKa = 3.44VEE131 pKa = 5.0VSTKK135 pKa = 10.15QDD137 pKa = 2.76GSLCITLAATTSSPTTPDD155 pKa = 2.92GLVQLVYY162 pKa = 10.8NLPGGLGSVEE172 pKa = 3.83IDD174 pKa = 3.43VASIEE179 pKa = 4.18FDD181 pKa = 3.17EE182 pKa = 5.64SAGTWTIIISGNLIITVPGLNWPSIEE208 pKa = 4.44LRR210 pKa = 11.84GLGIDD215 pKa = 3.45SKK217 pKa = 11.9GNISLQGGWINLPNQMALDD236 pKa = 4.39FYY238 pKa = 10.88GFSIALQALGFGSDD252 pKa = 5.24ASGKK256 pKa = 9.18WIGFNGDD263 pKa = 2.92INLVEE268 pKa = 4.34GLSLGGSVQGLKK280 pKa = 10.9VNLTTGSISFNGVGISFEE298 pKa = 4.07IPGVITIDD306 pKa = 3.57GEE308 pKa = 3.88IDD310 pKa = 4.07HH311 pKa = 6.76IHH313 pKa = 6.35VDD315 pKa = 3.6ANTPNDD321 pKa = 3.87LVPYY325 pKa = 10.1GLMPSIFNSIAPVGPTGPKK344 pKa = 10.34SIDD347 pKa = 3.3LFAGSVKK354 pKa = 10.54VQVLPDD360 pKa = 3.64EE361 pKa = 4.71MDD363 pKa = 3.95LEE365 pKa = 4.28VDD367 pKa = 3.41ANFIVGNFGGQSVFFLALDD386 pKa = 3.49VEE388 pKa = 4.84LPVGIPIFMDD398 pKa = 3.51VSLYY402 pKa = 10.87GLSGLVATGLDD413 pKa = 4.23PDD415 pKa = 4.87PEE417 pKa = 5.62PNDD420 pKa = 4.23TWWQWYY426 pKa = 9.19KK427 pKa = 11.2YY428 pKa = 9.17PAGTSGPQLNATPDD442 pKa = 3.42YY443 pKa = 10.88SATDD447 pKa = 3.67FTKK450 pKa = 10.28WLVPQQGAFAIGGGCTIGTSADD472 pKa = 3.36DD473 pKa = 4.91GYY475 pKa = 9.38TVSAAIMLVILMPGPVISLIGKK497 pKa = 9.87ANILSKK503 pKa = 10.47RR504 pKa = 11.84ISGADD509 pKa = 2.97QDD511 pKa = 4.92ANFEE515 pKa = 4.29AMATYY520 pKa = 10.38DD521 pKa = 3.98GNSGTFDD528 pKa = 3.45LTIDD532 pKa = 3.58AQYY535 pKa = 10.59SIPVVLDD542 pKa = 3.43IEE544 pKa = 4.56ATAEE548 pKa = 4.21LYY550 pKa = 10.96VDD552 pKa = 4.06ATAGEE557 pKa = 4.48WFFAMGKK564 pKa = 9.0PPHH567 pKa = 5.33TQRR570 pKa = 11.84VSARR574 pKa = 11.84IFDD577 pKa = 3.72IFEE580 pKa = 3.78TDD582 pKa = 2.71AYY584 pKa = 10.55FVVSNAGLITGTWTGYY600 pKa = 8.9QGSWSFGPLSASLDD614 pKa = 3.57AYY616 pKa = 10.85LATLAAIQWSPLQIAGGIEE635 pKa = 3.93LHH637 pKa = 7.09GSVQLSAFGIGVGISADD654 pKa = 3.59ALLEE658 pKa = 4.38GCAPNPFWVHH668 pKa = 5.99GEE670 pKa = 3.94LSVEE674 pKa = 5.31LDD676 pKa = 4.43LPWPLPNVGGTISLSWGGDD695 pKa = 3.47DD696 pKa = 5.37GSIPPVPLALSHH708 pKa = 7.03IDD710 pKa = 3.75AALSDD715 pKa = 4.33HH716 pKa = 6.84GDD718 pKa = 3.73SAGKK722 pKa = 8.64PASDD726 pKa = 3.55HH727 pKa = 5.97YY728 pKa = 11.69VLLAHH733 pKa = 7.43RR734 pKa = 11.84SNGPFPDD741 pKa = 3.62LTVQYY746 pKa = 10.21DD747 pKa = 3.49NPQAPGILDD756 pKa = 3.54LTGTALTNWQNRR768 pKa = 11.84VSSGNLADD776 pKa = 4.8MLPDD780 pKa = 3.45MTPDD784 pKa = 3.18NTSRR788 pKa = 11.84TQMAPVLPQDD798 pKa = 3.26THH800 pKa = 6.18FVLNFSHH807 pKa = 6.96PVVDD811 pKa = 3.89MSGGFNNALPQAQLPPEE828 pKa = 4.6VVTTPPPPPPTEE840 pKa = 3.87VGKK843 pKa = 10.61DD844 pKa = 3.29DD845 pKa = 4.09MSNINPNPPSVQFLIRR861 pKa = 11.84HH862 pKa = 5.09SLIDD866 pKa = 3.36VSLYY870 pKa = 10.42EE871 pKa = 4.18WVGGAWSLVASIPPVPPTEE890 pKa = 4.73ASAQADD896 pKa = 3.78TTYY899 pKa = 11.45LSGVWLAPTSATQIQLKK916 pKa = 8.58VVPWMMVEE924 pKa = 4.27GEE926 pKa = 4.31FWTVSWSSTSTPQNYY941 pKa = 7.4GTSFNNQGLDD951 pKa = 3.59FQTTNTQPAAIGTGNAPGLQQGLAFTNAGGSQQPSVAITFPQPEE995 pKa = 4.44VLSSLTAVVAVDD1007 pKa = 3.68EE1008 pKa = 5.04GEE1010 pKa = 4.42LLYY1013 pKa = 10.75AYY1015 pKa = 8.01NAPTCVGDD1023 pKa = 4.4GVTLTPTAQSWDD1035 pKa = 3.88SNFMQWTLTFDD1046 pKa = 3.91PEE1048 pKa = 4.06ATPISTLTLSMQNTSGATLVLYY1070 pKa = 10.7ALDD1073 pKa = 3.71YY1074 pKa = 8.63TVPPTAMAILPKK1086 pKa = 10.67APATYY1091 pKa = 10.87ALRR1094 pKa = 11.84VTTKK1098 pKa = 10.26IEE1100 pKa = 3.73AARR1103 pKa = 11.84VNGGTPQYY1111 pKa = 9.35QTAPDD1116 pKa = 4.21GSPIVEE1122 pKa = 4.22FAYY1125 pKa = 10.18FQTACGPGIAEE1136 pKa = 4.61IGASIPNTSGLPSTPTNTTPVPAEE1160 pKa = 3.76AANFPQLAVNCSTMQQPFTSFPLGGAIDD1188 pKa = 5.15DD1189 pKa = 4.16LTTYY1193 pKa = 7.25TQWSWPLNGAVTAYY1207 pKa = 10.33YY1208 pKa = 10.29GYY1210 pKa = 10.68DD1211 pKa = 3.41VNVEE1215 pKa = 4.09FCEE1218 pKa = 4.66SYY1220 pKa = 11.62VNALYY1225 pKa = 10.57VAAGGRR1231 pKa = 11.84GIANSLHH1238 pKa = 5.59FRR1240 pKa = 11.84CVDD1243 pKa = 3.27RR1244 pKa = 11.84NQNHH1248 pKa = 7.23LLLQTLAIHH1257 pKa = 6.1VPSIPQQSALVATALTVPLPSVIQPSDD1284 pKa = 3.37QDD1286 pKa = 3.61GLLTLTGAQLAGLEE1300 pKa = 4.0KK1301 pKa = 10.33RR1302 pKa = 11.84ALVKK1306 pKa = 9.99IDD1308 pKa = 4.82SNLQPDD1314 pKa = 4.39TAASLQNTAITNVSQLTGPGIQQLNPGIIGIILKK1348 pKa = 9.69EE1349 pKa = 4.03RR1350 pKa = 11.84QEE1352 pKa = 3.97QEE1354 pKa = 3.39AAATARR1360 pKa = 11.84QLWFQPLAPQMQYY1373 pKa = 9.79TLDD1376 pKa = 3.63VVAGSWLGATSDD1388 pKa = 4.36FYY1390 pKa = 10.93PAAGAGTGSLEE1401 pKa = 4.48EE1402 pKa = 4.47IFSASDD1408 pKa = 3.96AISLLKK1414 pKa = 10.67DD1415 pKa = 3.29LEE1417 pKa = 4.25NWYY1420 pKa = 8.14TAQAALTTLEE1430 pKa = 4.18RR1431 pKa = 11.84VTFTTSRR1438 pKa = 11.84YY1439 pKa = 8.54ATFSSQMSNAADD1451 pKa = 3.49QLAGVAGTAPLRR1463 pKa = 11.84HH1464 pKa = 6.33YY1465 pKa = 7.76NTSVDD1470 pKa = 3.77VNTWLADD1477 pKa = 3.48AANGYY1482 pKa = 9.14SAFTTAQTIYY1492 pKa = 8.17TQQRR1496 pKa = 11.84ATLAGVVAQFDD1507 pKa = 3.84PRR1509 pKa = 11.84ADD1511 pKa = 3.77DD1512 pKa = 4.39LAPGAPWPDD1521 pKa = 3.57NGNHH1525 pKa = 6.49ALVTARR1531 pKa = 11.84EE1532 pKa = 4.32ATSQAWQALQQTSVPVFDD1550 pKa = 5.05ALIGALGRR1558 pKa = 11.84ADD1560 pKa = 3.44LTSTQKK1566 pKa = 10.51PVAVPDD1572 pKa = 3.7TEE1574 pKa = 4.21MTLFLDD1580 pKa = 3.49PTGFDD1585 pKa = 2.87IRR1587 pKa = 11.84AILLEE1592 pKa = 4.41SPEE1595 pKa = 4.03ALPWQRR1601 pKa = 11.84IWQWIQLTPISPSSLGLKK1619 pKa = 10.12QIDD1622 pKa = 3.78VLWNSDD1628 pKa = 3.21NTRR1631 pKa = 11.84GLILPLGSPRR1641 pKa = 11.84GRR1643 pKa = 11.84YY1644 pKa = 6.25TLSITFQGNLGAEE1657 pKa = 4.43APCITLNGLPVTEE1670 pKa = 4.44AATIGVLSLGAVIHH1684 pKa = 6.43KK1685 pKa = 9.38PRR1687 pKa = 11.84NLL1689 pKa = 3.35
Molecular weight: 177.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q1SJK8|A0A4Q1SJK8_9BACT Uncharacterized protein OS=Acidipila dinghuensis OX=1560006 GN=ESZ00_07015 PE=4 SV=1
MM1 pKa = 8.0PKK3 pKa = 9.02RR4 pKa = 11.84TFQPNRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SKK15 pKa = 9.6VHH17 pKa = 5.91GFRR20 pKa = 11.84SRR22 pKa = 11.84MKK24 pKa = 8.69TKK26 pKa = 10.4SGAAVLSRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.64GRR40 pKa = 11.84KK41 pKa = 8.23RR42 pKa = 11.84VSVSASYY49 pKa = 11.04RR50 pKa = 11.84DD51 pKa = 3.38
MM1 pKa = 8.0PKK3 pKa = 9.02RR4 pKa = 11.84TFQPNRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SKK15 pKa = 9.6VHH17 pKa = 5.91GFRR20 pKa = 11.84SRR22 pKa = 11.84MKK24 pKa = 8.69TKK26 pKa = 10.4SGAAVLSRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.64GRR40 pKa = 11.84KK41 pKa = 8.23RR42 pKa = 11.84VSVSASYY49 pKa = 11.04RR50 pKa = 11.84DD51 pKa = 3.38
Molecular weight: 6.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1493901 |
29 |
2828 |
382.2 |
41.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.201 ± 0.051 | 0.811 ± 0.012 |
5.098 ± 0.031 | 5.661 ± 0.045 |
3.854 ± 0.028 | 8.031 ± 0.039 |
2.422 ± 0.02 | 4.873 ± 0.028 |
3.361 ± 0.034 | 10.094 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.191 ± 0.017 | 3.247 ± 0.037 |
5.344 ± 0.027 | 3.898 ± 0.029 |
6.206 ± 0.045 | 6.378 ± 0.038 |
5.989 ± 0.046 | 6.957 ± 0.029 |
1.463 ± 0.019 | 2.922 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
