
Spirochaeta thermophila (strain ATCC 700085 / DSM 6578 / Z-1203)
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Spirochaetales; Spirochaetaceae; Spirochaeta; Spirochaeta thermophila
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
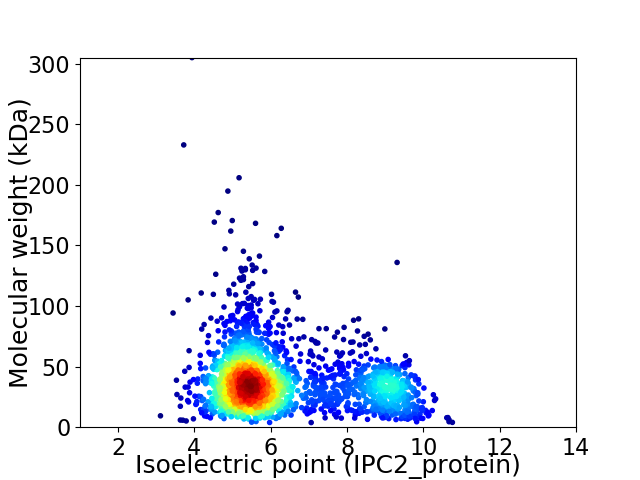
Virtual 2D-PAGE plot for 2249 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G0GBM5|G0GBM5_SPITZ Methyltransferase type 11 OS=Spirochaeta thermophila (strain ATCC 700085 / DSM 6578 / Z-1203) OX=869211 GN=Spith_0828 PE=4 SV=1
MM1 pKa = 7.47GFGSGTLFGQSAIPAQSGNYY21 pKa = 9.19LQVTGTPQNGDD32 pKa = 3.4EE33 pKa = 4.53DD34 pKa = 5.14PYY36 pKa = 10.8MVLFYY41 pKa = 10.01EE42 pKa = 4.57VPDD45 pKa = 4.7TITSTLYY52 pKa = 10.52FAVEE56 pKa = 4.16HH57 pKa = 6.96PGLSGAAPDD66 pKa = 3.8QGTGGTWEE74 pKa = 4.06FFLVGGAGALTQSRR88 pKa = 11.84MLNFANDD95 pKa = 3.86TEE97 pKa = 4.53VLAGTVLSTIQATNEE112 pKa = 4.11TGWVYY117 pKa = 10.8FDD119 pKa = 4.95GVLPSQGEE127 pKa = 4.5HH128 pKa = 5.73IGNRR132 pKa = 11.84YY133 pKa = 7.56YY134 pKa = 10.97FKK136 pKa = 10.6IVAKK140 pKa = 10.53APNGNKK146 pKa = 9.93NGFRR150 pKa = 11.84CDD152 pKa = 3.12VSYY155 pKa = 11.59SNSGTPTGDD164 pKa = 2.85SSIRR168 pKa = 11.84AFAYY172 pKa = 9.71VWTLALLDD180 pKa = 3.77NVGRR184 pKa = 11.84LWDD187 pKa = 4.84LYY189 pKa = 10.65PFVPDD194 pKa = 3.44SAGPTDD200 pKa = 3.87TIDD203 pKa = 3.45YY204 pKa = 10.03KK205 pKa = 11.15NWDD208 pKa = 3.74MDD210 pKa = 3.65GGADD214 pKa = 5.03TLAAWDD220 pKa = 3.86KK221 pKa = 11.33SGNPLTAPTPSGDD234 pKa = 3.3GATWPDD240 pKa = 3.7DD241 pKa = 3.64VATTSYY247 pKa = 10.86TIGSEE252 pKa = 4.65TNGTWHH258 pKa = 6.5LQITEE263 pKa = 4.31GNGGEE268 pKa = 4.08PAINTSLFWFTLNDD282 pKa = 3.51TTADD286 pKa = 3.72DD287 pKa = 3.7TDD289 pKa = 3.81VPLRR293 pKa = 11.84AYY295 pKa = 9.17SAPYY299 pKa = 8.72TPPAPDD305 pKa = 3.56HH306 pKa = 6.47VVISPDD312 pKa = 3.17AQTVQTGQTASLTLQIVDD330 pKa = 3.82ASGSPVPYY338 pKa = 10.17VRR340 pKa = 11.84NLYY343 pKa = 8.57VTVSGSATISPDD355 pKa = 3.53NNGTPQTEE363 pKa = 4.43LLTTSSDD370 pKa = 3.79GIATFTVTDD379 pKa = 4.91GSAEE383 pKa = 4.28TVTVTVYY390 pKa = 10.37WDD392 pKa = 3.44GTGGSDD398 pKa = 4.11SFGTSSYY405 pKa = 10.18ATATVDD411 pKa = 3.56VAASLPPVISSAANTTIPIGTGPYY435 pKa = 10.28ALADD439 pKa = 3.55VTVTDD444 pKa = 3.57VSGGQVTAANGIRR457 pKa = 11.84IRR459 pKa = 11.84LGGGLSAQFDD469 pKa = 3.99AAVVSLTTSGTAVSNGRR486 pKa = 11.84VANPVTPTYY495 pKa = 10.65EE496 pKa = 3.85SGNTVVFIPVLVDD509 pKa = 5.16FSAGEE514 pKa = 4.19SVTISGLALDD524 pKa = 4.75SATLSPSSGYY534 pKa = 10.91LEE536 pKa = 4.75LSVDD540 pKa = 4.77GGTTWVSVDD549 pKa = 3.68DD550 pKa = 4.47KK551 pKa = 11.67VIVLQGSSYY560 pKa = 11.01VWDD563 pKa = 4.66GDD565 pKa = 3.96TSTSWTDD572 pKa = 3.19GNNWQGGVAPPDD584 pKa = 4.2DD585 pKa = 3.94GTAEE589 pKa = 3.83ITLPAGCTYY598 pKa = 10.56YY599 pKa = 10.06PQLVSGEE606 pKa = 4.39SHH608 pKa = 5.76TVAQVVVTTGGSLTLDD624 pKa = 3.77AGSSLAVNGGLQDD637 pKa = 4.25DD638 pKa = 4.68GAVTVSGTLTVSGLLTVTGSLEE660 pKa = 4.13VQGGGSVSAGTVSSGGTIQSAGDD683 pKa = 3.24ITTGDD688 pKa = 3.67FTSTGSFTSTGVNTITASGDD708 pKa = 3.29VSISGTFSDD717 pKa = 4.69PSARR721 pKa = 11.84CTLTMSGPSTSLTSSQALGNLVLSGTASVGLGSSLSLAGGLSVPPGTTLDD771 pKa = 3.52AAGFDD776 pKa = 3.56VDD778 pKa = 3.77VAGTVTVGGMFDD790 pKa = 3.4ALGITLTVGGDD801 pKa = 3.44LSVTGTLTLTGATCVLDD818 pKa = 3.88GAGSQDD824 pKa = 3.38VTLGGAQPATLRR836 pKa = 11.84VANTGGVVRR845 pKa = 11.84FTDD848 pKa = 3.93AVTTDD853 pKa = 3.3TLQVVSAGSVTFEE866 pKa = 4.47GDD868 pKa = 3.42LTVNTSASLAAAGYY882 pKa = 10.36DD883 pKa = 3.39LAFNAGAGGQTTSFPGGITFPHH905 pKa = 6.51TGSLGLGNAAADD917 pKa = 3.76SFQAAGILTATACSSVQLGGTVSTAGAGGDD947 pKa = 3.77VSLGDD952 pKa = 3.84ADD954 pKa = 4.24TPVTLAADD962 pKa = 3.89TTVDD966 pKa = 3.26AGTGGVTLGGTVDD979 pKa = 3.38GGYY982 pKa = 10.64VLTVSAGGAVSFGGVVGGVTALAGLDD1008 pKa = 3.45ASGGSIVVGGDD1019 pKa = 3.23VVATGDD1025 pKa = 3.65VVMGGPVVLGADD1037 pKa = 3.46VVVDD1041 pKa = 3.68TSGGSGDD1048 pKa = 3.74VVFSSTVDD1056 pKa = 2.99GGYY1059 pKa = 10.59VLTVRR1064 pKa = 11.84AGGAVSFGGAVGGVTALAGLDD1085 pKa = 3.44ASGGSIAVGGDD1096 pKa = 3.37VVATGDD1102 pKa = 3.65VVMGGPVVLGADD1114 pKa = 3.46VVVDD1118 pKa = 3.68TSGGSGDD1125 pKa = 3.74VVFSSTVDD1133 pKa = 3.48GTWALTVQGGTGNIVFSGDD1152 pKa = 3.01IGGTTPLGALTIVSGAQVDD1171 pKa = 3.94LGGSVYY1177 pKa = 10.66TSDD1180 pKa = 4.55ADD1182 pKa = 4.11LSIAPPVRR1190 pKa = 11.84LTSAATDD1197 pKa = 3.14VWIEE1201 pKa = 4.02PGTGNITFGTTVEE1214 pKa = 4.4DD1215 pKa = 4.14SGSGAHH1221 pKa = 6.53NLILVGGGNLTFSGDD1236 pKa = 3.29VGATNEE1242 pKa = 4.03PAYY1245 pKa = 10.72LILDD1249 pKa = 3.51IAGDD1253 pKa = 3.62ITISGNVRR1261 pKa = 11.84AGNFVLYY1268 pKa = 10.65NGTVFLNGNTIEE1280 pKa = 4.36TTTGDD1285 pKa = 3.37VVLFGPAYY1293 pKa = 10.53GEE1295 pKa = 4.87DD1296 pKa = 4.01DD1297 pKa = 4.38PEE1299 pKa = 4.33TTPVDD1304 pKa = 3.62TNFRR1308 pKa = 11.84YY1309 pKa = 9.94PDD1311 pKa = 3.56MAGLAAVYY1319 pKa = 10.41DD1320 pKa = 4.19PGSYY1324 pKa = 10.75DD1325 pKa = 3.4GTFDD1329 pKa = 3.57AASLGGSTIDD1339 pKa = 3.59VGQNFYY1345 pKa = 11.73ANGVDD1350 pKa = 4.98LSASSAWTLDD1360 pKa = 3.09IPDD1363 pKa = 3.92NSASQRR1369 pKa = 11.84IAYY1372 pKa = 6.85PVPGGAYY1379 pKa = 7.36GTGSGTYY1386 pKa = 9.89AVAYY1390 pKa = 8.71NCTVAYY1396 pKa = 10.47SNAGPGVVSASTGNGNTDD1414 pKa = 2.97GGNNSGWDD1422 pKa = 3.7FTPPAIGATVEE1433 pKa = 4.25TLSDD1437 pKa = 3.54NVLHH1441 pKa = 5.57VTFSEE1446 pKa = 4.43DD1447 pKa = 3.35LEE1449 pKa = 4.38NSNGEE1454 pKa = 3.55ITAAVQAGLIRR1465 pKa = 11.84FHH1467 pKa = 7.43DD1468 pKa = 4.43GSGTYY1473 pKa = 9.83TFDD1476 pKa = 4.51EE1477 pKa = 5.3VYY1479 pKa = 10.7QDD1481 pKa = 3.83AACTIPLPPGDD1492 pKa = 3.57TDD1494 pKa = 3.7EE1495 pKa = 5.78FYY1497 pKa = 11.44LKK1499 pKa = 10.14ATGPNEE1505 pKa = 3.75AWNTDD1510 pKa = 2.95ATGADD1515 pKa = 3.95PGDD1518 pKa = 4.27ADD1520 pKa = 3.84STDD1523 pKa = 3.01TGGTHH1528 pKa = 5.7RR1529 pKa = 11.84TLVPRR1534 pKa = 11.84LYY1536 pKa = 9.96IEE1538 pKa = 4.41KK1539 pKa = 10.49GALFDD1544 pKa = 4.77AGKK1547 pKa = 10.42NPIGPYY1553 pKa = 9.62GINGGGPDD1561 pKa = 3.44TSAVDD1566 pKa = 3.36KK1567 pKa = 10.92APPVLYY1573 pKa = 10.13KK1574 pKa = 10.46VEE1576 pKa = 4.06YY1577 pKa = 10.61GRR1579 pKa = 11.84ADD1581 pKa = 3.32PAAAANTDD1589 pKa = 3.61YY1590 pKa = 10.99HH1591 pKa = 5.81GHH1593 pKa = 6.01NFFHH1597 pKa = 7.38LYY1599 pKa = 10.19YY1600 pKa = 9.99SEE1602 pKa = 4.02QVTIGTLGTGAANEE1616 pKa = 4.07RR1617 pKa = 11.84SEE1619 pKa = 5.76DD1620 pKa = 3.75SITNKK1625 pKa = 10.59GGDD1628 pKa = 3.45IEE1630 pKa = 6.06DD1631 pKa = 4.46DD1632 pKa = 3.94GGTHH1636 pKa = 6.73LVVEE1640 pKa = 5.73GYY1642 pKa = 10.48FSIDD1646 pKa = 3.01FGAPVSMEE1654 pKa = 3.74RR1655 pKa = 11.84GVRR1658 pKa = 11.84PGTPGGNSSNALYY1671 pKa = 8.19RR1672 pKa = 11.84TLPGLITSPDD1682 pKa = 3.44HH1683 pKa = 5.92EE1684 pKa = 4.46VVIFLSGYY1692 pKa = 10.17HH1693 pKa = 7.37DD1694 pKa = 3.69GTGWPGWHH1702 pKa = 6.55RR1703 pKa = 11.84NVPDD1707 pKa = 4.15PAGGTIDD1714 pKa = 3.73EE1715 pKa = 4.72VYY1717 pKa = 11.0DD1718 pKa = 3.88LGGQIEE1724 pKa = 4.94DD1725 pKa = 3.91NSPANNPLDD1734 pKa = 3.81TSVLPTFVLSSTGSFDD1750 pKa = 5.67LDD1752 pKa = 3.22WDD1754 pKa = 3.74VDD1756 pKa = 4.16PPALAPYY1763 pKa = 8.54TLAAGRR1769 pKa = 11.84YY1770 pKa = 8.18EE1771 pKa = 4.19GVTIDD1776 pKa = 3.79SDD1778 pKa = 3.61WNGRR1782 pKa = 11.84IDD1784 pKa = 5.23RR1785 pKa = 11.84IDD1787 pKa = 3.43FHH1789 pKa = 8.72FLDD1792 pKa = 5.41DD1793 pKa = 5.98GSAPWDD1799 pKa = 3.64SSTDD1803 pKa = 3.48HH1804 pKa = 7.58PDD1806 pKa = 3.17SQGGIRR1812 pKa = 11.84DD1813 pKa = 3.3ILLNYY1818 pKa = 9.73PGDD1821 pKa = 3.87SLNEE1825 pKa = 3.74LEE1827 pKa = 4.68AFMVGEE1833 pKa = 4.22TGGGSFVSTYY1843 pKa = 9.61NQAYY1847 pKa = 6.96GTGVDD1852 pKa = 3.3NSLFGGSISISDD1864 pKa = 4.07DD1865 pKa = 3.68PYY1867 pKa = 11.26FSLSLSSSAPWGLLTDD1883 pKa = 3.64ISVRR1887 pKa = 11.84YY1888 pKa = 9.47DD1889 pKa = 3.01HH1890 pKa = 6.82TKK1892 pKa = 10.76AYY1894 pKa = 9.08LTDD1897 pKa = 3.59LAGNLLEE1904 pKa = 6.19DD1905 pKa = 4.63IDD1907 pKa = 4.47PEE1909 pKa = 4.3VPIIEE1914 pKa = 4.2RR1915 pKa = 11.84VPPRR1919 pKa = 11.84IVFSLARR1926 pKa = 11.84AGDD1929 pKa = 3.69DD1930 pKa = 3.04KK1931 pKa = 11.71VYY1933 pKa = 10.95LRR1935 pKa = 11.84FSEE1938 pKa = 4.56PVFGDD1943 pKa = 3.53TAASVEE1949 pKa = 3.76IDD1951 pKa = 3.22AADD1954 pKa = 3.71FTVTGATGVLTVTGLQILTRR1974 pKa = 11.84WPANTTGAWEE1984 pKa = 4.51AYY1986 pKa = 10.12LLLAQPLTADD1996 pKa = 4.01DD1997 pKa = 4.22VVQGTVRR2004 pKa = 11.84ANANAVYY2011 pKa = 10.03DD2012 pKa = 4.15PSGNAMDD2019 pKa = 4.75PADD2022 pKa = 3.73IFRR2025 pKa = 11.84ITNVGLGVVSPLWATDD2041 pKa = 3.54GVHH2044 pKa = 6.63ADD2046 pKa = 3.67EE2047 pKa = 5.23VGGSSLRR2054 pKa = 11.84GFDD2057 pKa = 3.68GSGALLPRR2065 pKa = 11.84DD2066 pKa = 3.21ITLQARR2072 pKa = 11.84ILAPSISGDD2081 pKa = 3.5AMLLVYY2087 pKa = 10.51DD2088 pKa = 4.36VDD2090 pKa = 4.41PDD2092 pKa = 3.17ASYY2095 pKa = 12.11YY2096 pKa = 7.38MTTDD2100 pKa = 3.09EE2101 pKa = 4.43YY2102 pKa = 11.69NRR2104 pKa = 11.84TTFPDD2109 pKa = 3.18MWWPVSLPGLKK2120 pKa = 8.47NTANTEE2126 pKa = 3.8ARR2128 pKa = 11.84TVSPFSSQGALRR2140 pKa = 11.84TFLIPGTDD2148 pKa = 3.16PEE2150 pKa = 4.25MRR2152 pKa = 11.84EE2153 pKa = 4.05GGTVEE2158 pKa = 3.69FLFYY2162 pKa = 10.34IDD2164 pKa = 4.41NLYY2167 pKa = 10.36CARR2170 pKa = 11.84LTDD2173 pKa = 3.88EE2174 pKa = 5.37DD2175 pKa = 4.51DD2176 pKa = 4.02PRR2178 pKa = 11.84TIAPWSFGLSGLKK2191 pKa = 8.27EE2192 pKa = 3.69QRR2194 pKa = 11.84AGVTILNNVINPTQGEE2210 pKa = 4.63VTVVNYY2216 pKa = 10.19RR2217 pKa = 11.84LSRR2220 pKa = 11.84AGMVTIQVFALDD2232 pKa = 3.83GSLVRR2237 pKa = 11.84TLQRR2241 pKa = 11.84GVQGPGAYY2249 pKa = 8.15TVAWDD2254 pKa = 3.59GRR2256 pKa = 11.84NGSGRR2261 pKa = 11.84VVARR2265 pKa = 11.84GLYY2268 pKa = 9.33FIRR2271 pKa = 11.84VVGPGIDD2278 pKa = 3.59EE2279 pKa = 4.04YY2280 pKa = 11.29RR2281 pKa = 11.84KK2282 pKa = 10.45VMVVKK2287 pKa = 10.73
MM1 pKa = 7.47GFGSGTLFGQSAIPAQSGNYY21 pKa = 9.19LQVTGTPQNGDD32 pKa = 3.4EE33 pKa = 4.53DD34 pKa = 5.14PYY36 pKa = 10.8MVLFYY41 pKa = 10.01EE42 pKa = 4.57VPDD45 pKa = 4.7TITSTLYY52 pKa = 10.52FAVEE56 pKa = 4.16HH57 pKa = 6.96PGLSGAAPDD66 pKa = 3.8QGTGGTWEE74 pKa = 4.06FFLVGGAGALTQSRR88 pKa = 11.84MLNFANDD95 pKa = 3.86TEE97 pKa = 4.53VLAGTVLSTIQATNEE112 pKa = 4.11TGWVYY117 pKa = 10.8FDD119 pKa = 4.95GVLPSQGEE127 pKa = 4.5HH128 pKa = 5.73IGNRR132 pKa = 11.84YY133 pKa = 7.56YY134 pKa = 10.97FKK136 pKa = 10.6IVAKK140 pKa = 10.53APNGNKK146 pKa = 9.93NGFRR150 pKa = 11.84CDD152 pKa = 3.12VSYY155 pKa = 11.59SNSGTPTGDD164 pKa = 2.85SSIRR168 pKa = 11.84AFAYY172 pKa = 9.71VWTLALLDD180 pKa = 3.77NVGRR184 pKa = 11.84LWDD187 pKa = 4.84LYY189 pKa = 10.65PFVPDD194 pKa = 3.44SAGPTDD200 pKa = 3.87TIDD203 pKa = 3.45YY204 pKa = 10.03KK205 pKa = 11.15NWDD208 pKa = 3.74MDD210 pKa = 3.65GGADD214 pKa = 5.03TLAAWDD220 pKa = 3.86KK221 pKa = 11.33SGNPLTAPTPSGDD234 pKa = 3.3GATWPDD240 pKa = 3.7DD241 pKa = 3.64VATTSYY247 pKa = 10.86TIGSEE252 pKa = 4.65TNGTWHH258 pKa = 6.5LQITEE263 pKa = 4.31GNGGEE268 pKa = 4.08PAINTSLFWFTLNDD282 pKa = 3.51TTADD286 pKa = 3.72DD287 pKa = 3.7TDD289 pKa = 3.81VPLRR293 pKa = 11.84AYY295 pKa = 9.17SAPYY299 pKa = 8.72TPPAPDD305 pKa = 3.56HH306 pKa = 6.47VVISPDD312 pKa = 3.17AQTVQTGQTASLTLQIVDD330 pKa = 3.82ASGSPVPYY338 pKa = 10.17VRR340 pKa = 11.84NLYY343 pKa = 8.57VTVSGSATISPDD355 pKa = 3.53NNGTPQTEE363 pKa = 4.43LLTTSSDD370 pKa = 3.79GIATFTVTDD379 pKa = 4.91GSAEE383 pKa = 4.28TVTVTVYY390 pKa = 10.37WDD392 pKa = 3.44GTGGSDD398 pKa = 4.11SFGTSSYY405 pKa = 10.18ATATVDD411 pKa = 3.56VAASLPPVISSAANTTIPIGTGPYY435 pKa = 10.28ALADD439 pKa = 3.55VTVTDD444 pKa = 3.57VSGGQVTAANGIRR457 pKa = 11.84IRR459 pKa = 11.84LGGGLSAQFDD469 pKa = 3.99AAVVSLTTSGTAVSNGRR486 pKa = 11.84VANPVTPTYY495 pKa = 10.65EE496 pKa = 3.85SGNTVVFIPVLVDD509 pKa = 5.16FSAGEE514 pKa = 4.19SVTISGLALDD524 pKa = 4.75SATLSPSSGYY534 pKa = 10.91LEE536 pKa = 4.75LSVDD540 pKa = 4.77GGTTWVSVDD549 pKa = 3.68DD550 pKa = 4.47KK551 pKa = 11.67VIVLQGSSYY560 pKa = 11.01VWDD563 pKa = 4.66GDD565 pKa = 3.96TSTSWTDD572 pKa = 3.19GNNWQGGVAPPDD584 pKa = 4.2DD585 pKa = 3.94GTAEE589 pKa = 3.83ITLPAGCTYY598 pKa = 10.56YY599 pKa = 10.06PQLVSGEE606 pKa = 4.39SHH608 pKa = 5.76TVAQVVVTTGGSLTLDD624 pKa = 3.77AGSSLAVNGGLQDD637 pKa = 4.25DD638 pKa = 4.68GAVTVSGTLTVSGLLTVTGSLEE660 pKa = 4.13VQGGGSVSAGTVSSGGTIQSAGDD683 pKa = 3.24ITTGDD688 pKa = 3.67FTSTGSFTSTGVNTITASGDD708 pKa = 3.29VSISGTFSDD717 pKa = 4.69PSARR721 pKa = 11.84CTLTMSGPSTSLTSSQALGNLVLSGTASVGLGSSLSLAGGLSVPPGTTLDD771 pKa = 3.52AAGFDD776 pKa = 3.56VDD778 pKa = 3.77VAGTVTVGGMFDD790 pKa = 3.4ALGITLTVGGDD801 pKa = 3.44LSVTGTLTLTGATCVLDD818 pKa = 3.88GAGSQDD824 pKa = 3.38VTLGGAQPATLRR836 pKa = 11.84VANTGGVVRR845 pKa = 11.84FTDD848 pKa = 3.93AVTTDD853 pKa = 3.3TLQVVSAGSVTFEE866 pKa = 4.47GDD868 pKa = 3.42LTVNTSASLAAAGYY882 pKa = 10.36DD883 pKa = 3.39LAFNAGAGGQTTSFPGGITFPHH905 pKa = 6.51TGSLGLGNAAADD917 pKa = 3.76SFQAAGILTATACSSVQLGGTVSTAGAGGDD947 pKa = 3.77VSLGDD952 pKa = 3.84ADD954 pKa = 4.24TPVTLAADD962 pKa = 3.89TTVDD966 pKa = 3.26AGTGGVTLGGTVDD979 pKa = 3.38GGYY982 pKa = 10.64VLTVSAGGAVSFGGVVGGVTALAGLDD1008 pKa = 3.45ASGGSIVVGGDD1019 pKa = 3.23VVATGDD1025 pKa = 3.65VVMGGPVVLGADD1037 pKa = 3.46VVVDD1041 pKa = 3.68TSGGSGDD1048 pKa = 3.74VVFSSTVDD1056 pKa = 2.99GGYY1059 pKa = 10.59VLTVRR1064 pKa = 11.84AGGAVSFGGAVGGVTALAGLDD1085 pKa = 3.44ASGGSIAVGGDD1096 pKa = 3.37VVATGDD1102 pKa = 3.65VVMGGPVVLGADD1114 pKa = 3.46VVVDD1118 pKa = 3.68TSGGSGDD1125 pKa = 3.74VVFSSTVDD1133 pKa = 3.48GTWALTVQGGTGNIVFSGDD1152 pKa = 3.01IGGTTPLGALTIVSGAQVDD1171 pKa = 3.94LGGSVYY1177 pKa = 10.66TSDD1180 pKa = 4.55ADD1182 pKa = 4.11LSIAPPVRR1190 pKa = 11.84LTSAATDD1197 pKa = 3.14VWIEE1201 pKa = 4.02PGTGNITFGTTVEE1214 pKa = 4.4DD1215 pKa = 4.14SGSGAHH1221 pKa = 6.53NLILVGGGNLTFSGDD1236 pKa = 3.29VGATNEE1242 pKa = 4.03PAYY1245 pKa = 10.72LILDD1249 pKa = 3.51IAGDD1253 pKa = 3.62ITISGNVRR1261 pKa = 11.84AGNFVLYY1268 pKa = 10.65NGTVFLNGNTIEE1280 pKa = 4.36TTTGDD1285 pKa = 3.37VVLFGPAYY1293 pKa = 10.53GEE1295 pKa = 4.87DD1296 pKa = 4.01DD1297 pKa = 4.38PEE1299 pKa = 4.33TTPVDD1304 pKa = 3.62TNFRR1308 pKa = 11.84YY1309 pKa = 9.94PDD1311 pKa = 3.56MAGLAAVYY1319 pKa = 10.41DD1320 pKa = 4.19PGSYY1324 pKa = 10.75DD1325 pKa = 3.4GTFDD1329 pKa = 3.57AASLGGSTIDD1339 pKa = 3.59VGQNFYY1345 pKa = 11.73ANGVDD1350 pKa = 4.98LSASSAWTLDD1360 pKa = 3.09IPDD1363 pKa = 3.92NSASQRR1369 pKa = 11.84IAYY1372 pKa = 6.85PVPGGAYY1379 pKa = 7.36GTGSGTYY1386 pKa = 9.89AVAYY1390 pKa = 8.71NCTVAYY1396 pKa = 10.47SNAGPGVVSASTGNGNTDD1414 pKa = 2.97GGNNSGWDD1422 pKa = 3.7FTPPAIGATVEE1433 pKa = 4.25TLSDD1437 pKa = 3.54NVLHH1441 pKa = 5.57VTFSEE1446 pKa = 4.43DD1447 pKa = 3.35LEE1449 pKa = 4.38NSNGEE1454 pKa = 3.55ITAAVQAGLIRR1465 pKa = 11.84FHH1467 pKa = 7.43DD1468 pKa = 4.43GSGTYY1473 pKa = 9.83TFDD1476 pKa = 4.51EE1477 pKa = 5.3VYY1479 pKa = 10.7QDD1481 pKa = 3.83AACTIPLPPGDD1492 pKa = 3.57TDD1494 pKa = 3.7EE1495 pKa = 5.78FYY1497 pKa = 11.44LKK1499 pKa = 10.14ATGPNEE1505 pKa = 3.75AWNTDD1510 pKa = 2.95ATGADD1515 pKa = 3.95PGDD1518 pKa = 4.27ADD1520 pKa = 3.84STDD1523 pKa = 3.01TGGTHH1528 pKa = 5.7RR1529 pKa = 11.84TLVPRR1534 pKa = 11.84LYY1536 pKa = 9.96IEE1538 pKa = 4.41KK1539 pKa = 10.49GALFDD1544 pKa = 4.77AGKK1547 pKa = 10.42NPIGPYY1553 pKa = 9.62GINGGGPDD1561 pKa = 3.44TSAVDD1566 pKa = 3.36KK1567 pKa = 10.92APPVLYY1573 pKa = 10.13KK1574 pKa = 10.46VEE1576 pKa = 4.06YY1577 pKa = 10.61GRR1579 pKa = 11.84ADD1581 pKa = 3.32PAAAANTDD1589 pKa = 3.61YY1590 pKa = 10.99HH1591 pKa = 5.81GHH1593 pKa = 6.01NFFHH1597 pKa = 7.38LYY1599 pKa = 10.19YY1600 pKa = 9.99SEE1602 pKa = 4.02QVTIGTLGTGAANEE1616 pKa = 4.07RR1617 pKa = 11.84SEE1619 pKa = 5.76DD1620 pKa = 3.75SITNKK1625 pKa = 10.59GGDD1628 pKa = 3.45IEE1630 pKa = 6.06DD1631 pKa = 4.46DD1632 pKa = 3.94GGTHH1636 pKa = 6.73LVVEE1640 pKa = 5.73GYY1642 pKa = 10.48FSIDD1646 pKa = 3.01FGAPVSMEE1654 pKa = 3.74RR1655 pKa = 11.84GVRR1658 pKa = 11.84PGTPGGNSSNALYY1671 pKa = 8.19RR1672 pKa = 11.84TLPGLITSPDD1682 pKa = 3.44HH1683 pKa = 5.92EE1684 pKa = 4.46VVIFLSGYY1692 pKa = 10.17HH1693 pKa = 7.37DD1694 pKa = 3.69GTGWPGWHH1702 pKa = 6.55RR1703 pKa = 11.84NVPDD1707 pKa = 4.15PAGGTIDD1714 pKa = 3.73EE1715 pKa = 4.72VYY1717 pKa = 11.0DD1718 pKa = 3.88LGGQIEE1724 pKa = 4.94DD1725 pKa = 3.91NSPANNPLDD1734 pKa = 3.81TSVLPTFVLSSTGSFDD1750 pKa = 5.67LDD1752 pKa = 3.22WDD1754 pKa = 3.74VDD1756 pKa = 4.16PPALAPYY1763 pKa = 8.54TLAAGRR1769 pKa = 11.84YY1770 pKa = 8.18EE1771 pKa = 4.19GVTIDD1776 pKa = 3.79SDD1778 pKa = 3.61WNGRR1782 pKa = 11.84IDD1784 pKa = 5.23RR1785 pKa = 11.84IDD1787 pKa = 3.43FHH1789 pKa = 8.72FLDD1792 pKa = 5.41DD1793 pKa = 5.98GSAPWDD1799 pKa = 3.64SSTDD1803 pKa = 3.48HH1804 pKa = 7.58PDD1806 pKa = 3.17SQGGIRR1812 pKa = 11.84DD1813 pKa = 3.3ILLNYY1818 pKa = 9.73PGDD1821 pKa = 3.87SLNEE1825 pKa = 3.74LEE1827 pKa = 4.68AFMVGEE1833 pKa = 4.22TGGGSFVSTYY1843 pKa = 9.61NQAYY1847 pKa = 6.96GTGVDD1852 pKa = 3.3NSLFGGSISISDD1864 pKa = 4.07DD1865 pKa = 3.68PYY1867 pKa = 11.26FSLSLSSSAPWGLLTDD1883 pKa = 3.64ISVRR1887 pKa = 11.84YY1888 pKa = 9.47DD1889 pKa = 3.01HH1890 pKa = 6.82TKK1892 pKa = 10.76AYY1894 pKa = 9.08LTDD1897 pKa = 3.59LAGNLLEE1904 pKa = 6.19DD1905 pKa = 4.63IDD1907 pKa = 4.47PEE1909 pKa = 4.3VPIIEE1914 pKa = 4.2RR1915 pKa = 11.84VPPRR1919 pKa = 11.84IVFSLARR1926 pKa = 11.84AGDD1929 pKa = 3.69DD1930 pKa = 3.04KK1931 pKa = 11.71VYY1933 pKa = 10.95LRR1935 pKa = 11.84FSEE1938 pKa = 4.56PVFGDD1943 pKa = 3.53TAASVEE1949 pKa = 3.76IDD1951 pKa = 3.22AADD1954 pKa = 3.71FTVTGATGVLTVTGLQILTRR1974 pKa = 11.84WPANTTGAWEE1984 pKa = 4.51AYY1986 pKa = 10.12LLLAQPLTADD1996 pKa = 4.01DD1997 pKa = 4.22VVQGTVRR2004 pKa = 11.84ANANAVYY2011 pKa = 10.03DD2012 pKa = 4.15PSGNAMDD2019 pKa = 4.75PADD2022 pKa = 3.73IFRR2025 pKa = 11.84ITNVGLGVVSPLWATDD2041 pKa = 3.54GVHH2044 pKa = 6.63ADD2046 pKa = 3.67EE2047 pKa = 5.23VGGSSLRR2054 pKa = 11.84GFDD2057 pKa = 3.68GSGALLPRR2065 pKa = 11.84DD2066 pKa = 3.21ITLQARR2072 pKa = 11.84ILAPSISGDD2081 pKa = 3.5AMLLVYY2087 pKa = 10.51DD2088 pKa = 4.36VDD2090 pKa = 4.41PDD2092 pKa = 3.17ASYY2095 pKa = 12.11YY2096 pKa = 7.38MTTDD2100 pKa = 3.09EE2101 pKa = 4.43YY2102 pKa = 11.69NRR2104 pKa = 11.84TTFPDD2109 pKa = 3.18MWWPVSLPGLKK2120 pKa = 8.47NTANTEE2126 pKa = 3.8ARR2128 pKa = 11.84TVSPFSSQGALRR2140 pKa = 11.84TFLIPGTDD2148 pKa = 3.16PEE2150 pKa = 4.25MRR2152 pKa = 11.84EE2153 pKa = 4.05GGTVEE2158 pKa = 3.69FLFYY2162 pKa = 10.34IDD2164 pKa = 4.41NLYY2167 pKa = 10.36CARR2170 pKa = 11.84LTDD2173 pKa = 3.88EE2174 pKa = 5.37DD2175 pKa = 4.51DD2176 pKa = 4.02PRR2178 pKa = 11.84TIAPWSFGLSGLKK2191 pKa = 8.27EE2192 pKa = 3.69QRR2194 pKa = 11.84AGVTILNNVINPTQGEE2210 pKa = 4.63VTVVNYY2216 pKa = 10.19RR2217 pKa = 11.84LSRR2220 pKa = 11.84AGMVTIQVFALDD2232 pKa = 3.83GSLVRR2237 pKa = 11.84TLQRR2241 pKa = 11.84GVQGPGAYY2249 pKa = 8.15TVAWDD2254 pKa = 3.59GRR2256 pKa = 11.84NGSGRR2261 pKa = 11.84VVARR2265 pKa = 11.84GLYY2268 pKa = 9.33FIRR2271 pKa = 11.84VVGPGIDD2278 pKa = 3.59EE2279 pKa = 4.04YY2280 pKa = 11.29RR2281 pKa = 11.84KK2282 pKa = 10.45VMVVKK2287 pKa = 10.73
Molecular weight: 232.97 kDa
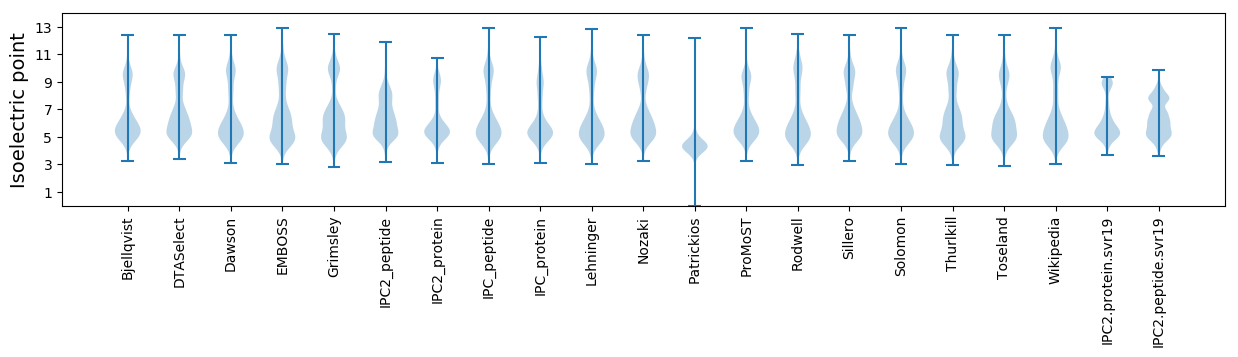
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G0GE59|G0GE59_SPITZ Permease YjgP/YjgQ family protein OS=Spirochaeta thermophila (strain ATCC 700085 / DSM 6578 / Z-1203) OX=869211 GN=Spith_1140 PE=4 SV=1
MM1 pKa = 7.6ICTTLSRR8 pKa = 11.84LLGFVRR14 pKa = 11.84VGVVAAVFGASGKK27 pKa = 10.63ADD29 pKa = 3.31VLNAVFNIPNNLRR42 pKa = 11.84KK43 pKa = 10.08LMAEE47 pKa = 4.33GALSSAFIPVLTQTHH62 pKa = 4.84QQDD65 pKa = 3.17PSGRR69 pKa = 11.84VSRR72 pKa = 11.84RR73 pKa = 11.84LMSTILGFQIIVLVPLIAAGIAGARR98 pKa = 11.84TIVPVLLDD106 pKa = 3.83FPDD109 pKa = 4.48PGKK112 pKa = 9.56MALSISLFRR121 pKa = 11.84WFLPYY126 pKa = 10.26TFLVSISAVLMGTLNSHH143 pKa = 6.28HH144 pKa = 7.03RR145 pKa = 11.84FFIPAVTPLLFSLSVIGCILLAGNRR170 pKa = 11.84LDD172 pKa = 3.87VYY174 pKa = 11.68AMALGVLIGGMMQILFQIPSILRR197 pKa = 11.84RR198 pKa = 11.84GYY200 pKa = 10.11SLIPNLHH207 pKa = 5.89FHH209 pKa = 7.4DD210 pKa = 4.65PPFRR214 pKa = 11.84EE215 pKa = 4.44VLRR218 pKa = 11.84LWGPIVASSSLLVIDD233 pKa = 3.68QQVAILFASGLEE245 pKa = 4.33DD246 pKa = 3.81GSTSALTNAIVFWQLPFGIFSASINTAFFPRR277 pKa = 11.84FAQDD281 pKa = 3.09ALDD284 pKa = 3.74RR285 pKa = 11.84TKK287 pKa = 10.26TALRR291 pKa = 11.84SSVEE295 pKa = 3.9QGLLALGFFLLPASLAMGILSHH317 pKa = 7.31DD318 pKa = 4.42IISAALQRR326 pKa = 11.84GAFLSSHH333 pKa = 5.97TEE335 pKa = 3.31MTAQVLRR342 pKa = 11.84AYY344 pKa = 10.21LPGMFFVGSFNLLQRR359 pKa = 11.84VSYY362 pKa = 11.0SKK364 pKa = 11.34GEE366 pKa = 4.14SVKK369 pKa = 10.7PFILVFLVVALDD381 pKa = 3.54ILLSFILKK389 pKa = 7.34EE390 pKa = 3.87TSLRR394 pKa = 11.84VAGLALAHH402 pKa = 6.3SLSFFTGSILLFLLTARR419 pKa = 11.84EE420 pKa = 4.22LEE422 pKa = 4.68GFPSPQFTRR431 pKa = 11.84EE432 pKa = 3.84LGKK435 pKa = 9.61MVAALIPFGIVLWGVRR451 pKa = 11.84VFLGDD456 pKa = 3.08RR457 pKa = 11.84WRR459 pKa = 11.84EE460 pKa = 3.79GSSFSYY466 pKa = 10.45FLLVGLLIIGSAVLILLMYY485 pKa = 10.75RR486 pKa = 11.84LFRR489 pKa = 11.84VDD491 pKa = 3.44LSKK494 pKa = 10.91RR495 pKa = 11.84IPLRR499 pKa = 11.84RR500 pKa = 11.84NRR502 pKa = 3.71
MM1 pKa = 7.6ICTTLSRR8 pKa = 11.84LLGFVRR14 pKa = 11.84VGVVAAVFGASGKK27 pKa = 10.63ADD29 pKa = 3.31VLNAVFNIPNNLRR42 pKa = 11.84KK43 pKa = 10.08LMAEE47 pKa = 4.33GALSSAFIPVLTQTHH62 pKa = 4.84QQDD65 pKa = 3.17PSGRR69 pKa = 11.84VSRR72 pKa = 11.84RR73 pKa = 11.84LMSTILGFQIIVLVPLIAAGIAGARR98 pKa = 11.84TIVPVLLDD106 pKa = 3.83FPDD109 pKa = 4.48PGKK112 pKa = 9.56MALSISLFRR121 pKa = 11.84WFLPYY126 pKa = 10.26TFLVSISAVLMGTLNSHH143 pKa = 6.28HH144 pKa = 7.03RR145 pKa = 11.84FFIPAVTPLLFSLSVIGCILLAGNRR170 pKa = 11.84LDD172 pKa = 3.87VYY174 pKa = 11.68AMALGVLIGGMMQILFQIPSILRR197 pKa = 11.84RR198 pKa = 11.84GYY200 pKa = 10.11SLIPNLHH207 pKa = 5.89FHH209 pKa = 7.4DD210 pKa = 4.65PPFRR214 pKa = 11.84EE215 pKa = 4.44VLRR218 pKa = 11.84LWGPIVASSSLLVIDD233 pKa = 3.68QQVAILFASGLEE245 pKa = 4.33DD246 pKa = 3.81GSTSALTNAIVFWQLPFGIFSASINTAFFPRR277 pKa = 11.84FAQDD281 pKa = 3.09ALDD284 pKa = 3.74RR285 pKa = 11.84TKK287 pKa = 10.26TALRR291 pKa = 11.84SSVEE295 pKa = 3.9QGLLALGFFLLPASLAMGILSHH317 pKa = 7.31DD318 pKa = 4.42IISAALQRR326 pKa = 11.84GAFLSSHH333 pKa = 5.97TEE335 pKa = 3.31MTAQVLRR342 pKa = 11.84AYY344 pKa = 10.21LPGMFFVGSFNLLQRR359 pKa = 11.84VSYY362 pKa = 11.0SKK364 pKa = 11.34GEE366 pKa = 4.14SVKK369 pKa = 10.7PFILVFLVVALDD381 pKa = 3.54ILLSFILKK389 pKa = 7.34EE390 pKa = 3.87TSLRR394 pKa = 11.84VAGLALAHH402 pKa = 6.3SLSFFTGSILLFLLTARR419 pKa = 11.84EE420 pKa = 4.22LEE422 pKa = 4.68GFPSPQFTRR431 pKa = 11.84EE432 pKa = 3.84LGKK435 pKa = 9.61MVAALIPFGIVLWGVRR451 pKa = 11.84VFLGDD456 pKa = 3.08RR457 pKa = 11.84WRR459 pKa = 11.84EE460 pKa = 3.79GSSFSYY466 pKa = 10.45FLLVGLLIIGSAVLILLMYY485 pKa = 10.75RR486 pKa = 11.84LFRR489 pKa = 11.84VDD491 pKa = 3.44LSKK494 pKa = 10.91RR495 pKa = 11.84IPLRR499 pKa = 11.84RR500 pKa = 11.84NRR502 pKa = 3.71
Molecular weight: 54.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
788271 |
31 |
2848 |
350.5 |
39.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.142 ± 0.052 | 0.821 ± 0.019 |
4.755 ± 0.041 | 8.365 ± 0.072 |
4.183 ± 0.031 | 8.188 ± 0.062 |
2.18 ± 0.029 | 5.231 ± 0.044 |
3.749 ± 0.045 | 11.178 ± 0.073 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.139 ± 0.022 | 2.091 ± 0.03 |
5.182 ± 0.045 | 2.62 ± 0.029 |
7.667 ± 0.075 | 5.537 ± 0.042 |
4.952 ± 0.062 | 8.344 ± 0.057 |
1.423 ± 0.025 | 3.252 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
