
Mangshi virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Seadornavirus; unclassified Seadornavirus
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
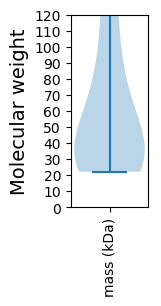
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A0S3JNF8|A0A0S3JNF8_9REOV VP12 OS=Mangshi virus OX=1766829 PE=4 SV=1
MM1 pKa = 7.4SRR3 pKa = 11.84NRR5 pKa = 11.84QIKK8 pKa = 9.23LRR10 pKa = 11.84NGQGTIIDD18 pKa = 3.75RR19 pKa = 11.84NGKK22 pKa = 9.86FEE24 pKa = 4.21VLVRR28 pKa = 11.84RR29 pKa = 11.84LDD31 pKa = 3.47IDD33 pKa = 3.67HH34 pKa = 7.04NLPLASTVSSGSRR47 pKa = 11.84VDD49 pKa = 3.15IVKK52 pKa = 10.23FLVDD56 pKa = 4.24FVGVTYY62 pKa = 10.13IDD64 pKa = 3.35SGGSVDD70 pKa = 4.42FSEE73 pKa = 6.21SEE75 pKa = 4.06LEE77 pKa = 4.18HH78 pKa = 6.35YY79 pKa = 9.95KK80 pKa = 11.15GNSTLLVASTAFGPLFIKK98 pKa = 10.44KK99 pKa = 9.15IRR101 pKa = 11.84QIPICNCAFIDD112 pKa = 3.59NKK114 pKa = 10.9YY115 pKa = 10.78NILAACRR122 pKa = 11.84GVLKK126 pKa = 9.5LTNDD130 pKa = 3.49RR131 pKa = 11.84NGYY134 pKa = 8.24RR135 pKa = 11.84YY136 pKa = 10.8GMILEE141 pKa = 4.63RR142 pKa = 11.84CHH144 pKa = 6.09PPKK147 pKa = 10.56INFTNLVEE155 pKa = 4.36AVVNLAYY162 pKa = 9.48MHH164 pKa = 6.74KK165 pKa = 9.86SEE167 pKa = 4.0PHH169 pKa = 6.09ALHH172 pKa = 7.09GDD174 pKa = 3.81VNPSNIMSDD183 pKa = 3.31QQGVLKK189 pKa = 10.74LVDD192 pKa = 3.84PVCILEE198 pKa = 4.22GQVNIANVDD207 pKa = 3.81YY208 pKa = 11.5EE209 pKa = 4.32EE210 pKa = 4.51LTQQEE215 pKa = 4.39EE216 pKa = 4.03MRR218 pKa = 11.84LFILSLLQILGSQFKK233 pKa = 10.73VRR235 pKa = 11.84IDD237 pKa = 3.43KK238 pKa = 11.18VKK240 pKa = 10.24IDD242 pKa = 3.7YY243 pKa = 9.71SKK245 pKa = 10.72CQPEE249 pKa = 4.2FTISDD254 pKa = 3.76DD255 pKa = 3.75SSNPSLADD263 pKa = 3.14VLSFNVSDD271 pKa = 4.54AIEE274 pKa = 4.13WKK276 pKa = 10.65DD277 pKa = 3.54RR278 pKa = 11.84MLSPRR283 pKa = 11.84AIPEE287 pKa = 3.44PSFRR291 pKa = 11.84HH292 pKa = 6.67DD293 pKa = 3.65YY294 pKa = 11.25YY295 pKa = 11.61KK296 pKa = 10.21LTEE299 pKa = 4.32LNEE302 pKa = 4.2QVSDD306 pKa = 5.06LDD308 pKa = 5.05DD309 pKa = 5.92DD310 pKa = 5.62DD311 pKa = 6.65DD312 pKa = 3.76II313 pKa = 6.96
MM1 pKa = 7.4SRR3 pKa = 11.84NRR5 pKa = 11.84QIKK8 pKa = 9.23LRR10 pKa = 11.84NGQGTIIDD18 pKa = 3.75RR19 pKa = 11.84NGKK22 pKa = 9.86FEE24 pKa = 4.21VLVRR28 pKa = 11.84RR29 pKa = 11.84LDD31 pKa = 3.47IDD33 pKa = 3.67HH34 pKa = 7.04NLPLASTVSSGSRR47 pKa = 11.84VDD49 pKa = 3.15IVKK52 pKa = 10.23FLVDD56 pKa = 4.24FVGVTYY62 pKa = 10.13IDD64 pKa = 3.35SGGSVDD70 pKa = 4.42FSEE73 pKa = 6.21SEE75 pKa = 4.06LEE77 pKa = 4.18HH78 pKa = 6.35YY79 pKa = 9.95KK80 pKa = 11.15GNSTLLVASTAFGPLFIKK98 pKa = 10.44KK99 pKa = 9.15IRR101 pKa = 11.84QIPICNCAFIDD112 pKa = 3.59NKK114 pKa = 10.9YY115 pKa = 10.78NILAACRR122 pKa = 11.84GVLKK126 pKa = 9.5LTNDD130 pKa = 3.49RR131 pKa = 11.84NGYY134 pKa = 8.24RR135 pKa = 11.84YY136 pKa = 10.8GMILEE141 pKa = 4.63RR142 pKa = 11.84CHH144 pKa = 6.09PPKK147 pKa = 10.56INFTNLVEE155 pKa = 4.36AVVNLAYY162 pKa = 9.48MHH164 pKa = 6.74KK165 pKa = 9.86SEE167 pKa = 4.0PHH169 pKa = 6.09ALHH172 pKa = 7.09GDD174 pKa = 3.81VNPSNIMSDD183 pKa = 3.31QQGVLKK189 pKa = 10.74LVDD192 pKa = 3.84PVCILEE198 pKa = 4.22GQVNIANVDD207 pKa = 3.81YY208 pKa = 11.5EE209 pKa = 4.32EE210 pKa = 4.51LTQQEE215 pKa = 4.39EE216 pKa = 4.03MRR218 pKa = 11.84LFILSLLQILGSQFKK233 pKa = 10.73VRR235 pKa = 11.84IDD237 pKa = 3.43KK238 pKa = 11.18VKK240 pKa = 10.24IDD242 pKa = 3.7YY243 pKa = 9.71SKK245 pKa = 10.72CQPEE249 pKa = 4.2FTISDD254 pKa = 3.76DD255 pKa = 3.75SSNPSLADD263 pKa = 3.14VLSFNVSDD271 pKa = 4.54AIEE274 pKa = 4.13WKK276 pKa = 10.65DD277 pKa = 3.54RR278 pKa = 11.84MLSPRR283 pKa = 11.84AIPEE287 pKa = 3.44PSFRR291 pKa = 11.84HH292 pKa = 6.67DD293 pKa = 3.65YY294 pKa = 11.25YY295 pKa = 11.61KK296 pKa = 10.21LTEE299 pKa = 4.32LNEE302 pKa = 4.2QVSDD306 pKa = 5.06LDD308 pKa = 5.05DD309 pKa = 5.92DD310 pKa = 5.62DD311 pKa = 6.65DD312 pKa = 3.76II313 pKa = 6.96
Molecular weight: 35.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S3JNE2|A0A0S3JNE2_9REOV VP11 OS=Mangshi virus OX=1766829 PE=4 SV=1
MM1 pKa = 7.94DD2 pKa = 4.01VLSKK6 pKa = 10.91KK7 pKa = 8.32SLRR10 pKa = 11.84EE11 pKa = 3.54FLAFLQEE18 pKa = 4.43TPFEE22 pKa = 4.4NALSMTDD29 pKa = 3.02GTAPHH34 pKa = 6.74IPVNVVVQEE43 pKa = 4.1YY44 pKa = 10.83NRR46 pKa = 11.84DD47 pKa = 3.47PYY49 pKa = 10.89IEE51 pKa = 4.44GTTLLDD57 pKa = 3.75GAPRR61 pKa = 11.84TSVRR65 pKa = 11.84NVTGMIRR72 pKa = 11.84SILSYY77 pKa = 7.56VTCGSGNYY85 pKa = 10.03VGMVDD90 pKa = 3.28VYY92 pKa = 10.83TRR94 pKa = 11.84TRR96 pKa = 11.84SIFDD100 pKa = 3.25ILNSRR105 pKa = 11.84FVNVNTAGLNYY116 pKa = 9.45VRR118 pKa = 11.84LVGAMTVNKK127 pKa = 9.83MSVEE131 pKa = 3.67QRR133 pKa = 11.84KK134 pKa = 8.62RR135 pKa = 11.84VIICIKK141 pKa = 10.58LIAEE145 pKa = 4.48KK146 pKa = 10.92DD147 pKa = 3.94NNLDD151 pKa = 3.41AYY153 pKa = 11.67NNFVNNDD160 pKa = 3.28PFLRR164 pKa = 11.84ICEE167 pKa = 4.27VKK169 pKa = 10.79FNVHH173 pKa = 4.96DD174 pKa = 4.2HH175 pKa = 6.45KK176 pKa = 11.39NITSVPDD183 pKa = 3.52NLSVLGLAGLNKK195 pKa = 9.47YY196 pKa = 10.59LKK198 pKa = 9.85IEE200 pKa = 3.93RR201 pKa = 11.84LMVHH205 pKa = 6.07NGVRR209 pKa = 11.84SVNRR213 pKa = 11.84EE214 pKa = 3.66YY215 pKa = 11.18EE216 pKa = 4.08EE217 pKa = 4.24YY218 pKa = 8.83KK219 pKa = 9.96TLRR222 pKa = 11.84NGSAFRR228 pKa = 11.84LQITDD233 pKa = 3.43LQDD236 pKa = 3.27AYY238 pKa = 11.28RR239 pKa = 11.84DD240 pKa = 3.77ALNPAGGGRR249 pKa = 11.84PPIIVV254 pKa = 3.26
MM1 pKa = 7.94DD2 pKa = 4.01VLSKK6 pKa = 10.91KK7 pKa = 8.32SLRR10 pKa = 11.84EE11 pKa = 3.54FLAFLQEE18 pKa = 4.43TPFEE22 pKa = 4.4NALSMTDD29 pKa = 3.02GTAPHH34 pKa = 6.74IPVNVVVQEE43 pKa = 4.1YY44 pKa = 10.83NRR46 pKa = 11.84DD47 pKa = 3.47PYY49 pKa = 10.89IEE51 pKa = 4.44GTTLLDD57 pKa = 3.75GAPRR61 pKa = 11.84TSVRR65 pKa = 11.84NVTGMIRR72 pKa = 11.84SILSYY77 pKa = 7.56VTCGSGNYY85 pKa = 10.03VGMVDD90 pKa = 3.28VYY92 pKa = 10.83TRR94 pKa = 11.84TRR96 pKa = 11.84SIFDD100 pKa = 3.25ILNSRR105 pKa = 11.84FVNVNTAGLNYY116 pKa = 9.45VRR118 pKa = 11.84LVGAMTVNKK127 pKa = 9.83MSVEE131 pKa = 3.67QRR133 pKa = 11.84KK134 pKa = 8.62RR135 pKa = 11.84VIICIKK141 pKa = 10.58LIAEE145 pKa = 4.48KK146 pKa = 10.92DD147 pKa = 3.94NNLDD151 pKa = 3.41AYY153 pKa = 11.67NNFVNNDD160 pKa = 3.28PFLRR164 pKa = 11.84ICEE167 pKa = 4.27VKK169 pKa = 10.79FNVHH173 pKa = 4.96DD174 pKa = 4.2HH175 pKa = 6.45KK176 pKa = 11.39NITSVPDD183 pKa = 3.52NLSVLGLAGLNKK195 pKa = 9.47YY196 pKa = 10.59LKK198 pKa = 9.85IEE200 pKa = 3.93RR201 pKa = 11.84LMVHH205 pKa = 6.07NGVRR209 pKa = 11.84SVNRR213 pKa = 11.84EE214 pKa = 3.66YY215 pKa = 11.18EE216 pKa = 4.08EE217 pKa = 4.24YY218 pKa = 8.83KK219 pKa = 9.96TLRR222 pKa = 11.84NGSAFRR228 pKa = 11.84LQITDD233 pKa = 3.43LQDD236 pKa = 3.27AYY238 pKa = 11.28RR239 pKa = 11.84DD240 pKa = 3.77ALNPAGGGRR249 pKa = 11.84PPIIVV254 pKa = 3.26
Molecular weight: 28.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5983 |
199 |
1214 |
498.6 |
55.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.521 ± 0.642 | 1.086 ± 0.125 |
6.351 ± 0.282 | 5.081 ± 0.246 |
3.761 ± 0.226 | 5.833 ± 0.242 |
2.022 ± 0.208 | 6.251 ± 0.221 |
5.382 ± 0.496 | 8.524 ± 0.259 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.226 ± 0.225 | 5.649 ± 0.39 |
4.112 ± 0.357 | 3.777 ± 0.416 |
5.666 ± 0.242 | 7.12 ± 0.288 |
6.569 ± 0.495 | 7.672 ± 0.403 |
0.635 ± 0.143 | 3.727 ± 0.208 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
