
Wenzhou tombus-like virus 17
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
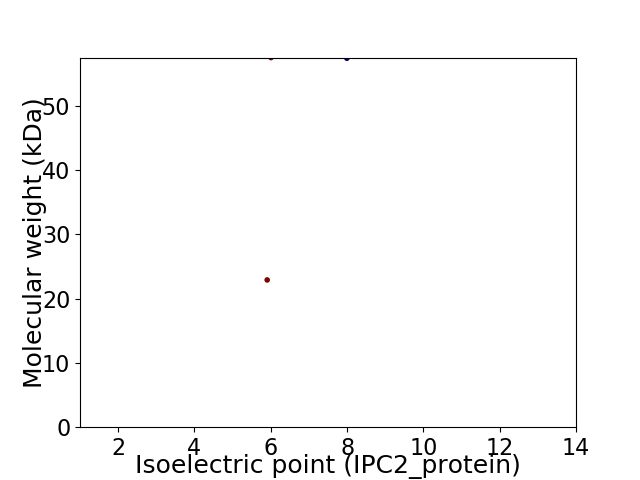
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
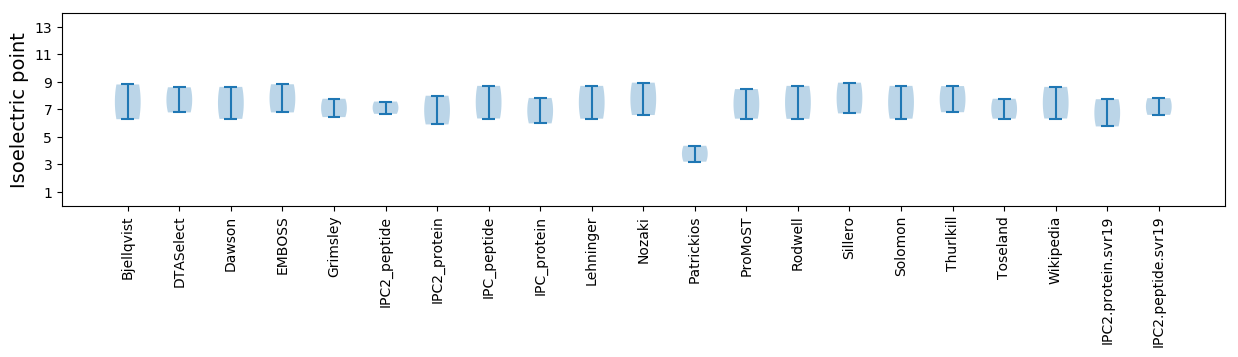
Protein with the lowest isoelectric point:
>tr|A0A1L3KH10|A0A1L3KH10_9VIRU RdRp catalytic domain-containing protein OS=Wenzhou tombus-like virus 17 OX=1923670 PE=4 SV=1
MM1 pKa = 7.56YY2 pKa = 9.38DD3 pKa = 3.08AQQPHH8 pKa = 5.75VLYY11 pKa = 10.16DD12 pKa = 3.45YY13 pKa = 10.17TGTGRR18 pKa = 11.84VYY20 pKa = 9.77IQSNFMVAFKK30 pKa = 10.3TGMCILVFGIMPILLADD47 pKa = 4.36YY48 pKa = 10.18ICPGYY53 pKa = 9.0NTVRR57 pKa = 11.84SYY59 pKa = 9.05QHH61 pKa = 5.93FWTFIKK67 pKa = 10.99YY68 pKa = 10.43VDD70 pKa = 3.42TDD72 pKa = 3.94EE73 pKa = 4.25IDD75 pKa = 3.25RR76 pKa = 11.84RR77 pKa = 11.84VDD79 pKa = 3.05INSASSLKK87 pKa = 9.46HH88 pKa = 5.7TDD90 pKa = 3.25PFLAWFRR97 pKa = 11.84YY98 pKa = 7.77EE99 pKa = 4.5RR100 pKa = 11.84NDD102 pKa = 3.2NGEE105 pKa = 4.35FYY107 pKa = 10.26PARR110 pKa = 11.84DD111 pKa = 3.58MLVSMEE117 pKa = 4.96LLAQCSHH124 pKa = 6.06AMQHH128 pKa = 5.7VLSDD132 pKa = 3.57DD133 pKa = 3.36KK134 pKa = 11.47TVFEE138 pKa = 5.22RR139 pKa = 11.84ILRR142 pKa = 11.84FCSTNCTVNLDD153 pKa = 3.84RR154 pKa = 11.84YY155 pKa = 10.54DD156 pKa = 3.68FLINGPRR163 pKa = 11.84QSIIHH168 pKa = 5.44NTAILSFSLYY178 pKa = 8.91KK179 pKa = 10.04QRR181 pKa = 11.84RR182 pKa = 11.84EE183 pKa = 3.88YY184 pKa = 11.01LKK186 pKa = 10.51EE187 pKa = 3.94FPFPQLQQ194 pKa = 3.1
MM1 pKa = 7.56YY2 pKa = 9.38DD3 pKa = 3.08AQQPHH8 pKa = 5.75VLYY11 pKa = 10.16DD12 pKa = 3.45YY13 pKa = 10.17TGTGRR18 pKa = 11.84VYY20 pKa = 9.77IQSNFMVAFKK30 pKa = 10.3TGMCILVFGIMPILLADD47 pKa = 4.36YY48 pKa = 10.18ICPGYY53 pKa = 9.0NTVRR57 pKa = 11.84SYY59 pKa = 9.05QHH61 pKa = 5.93FWTFIKK67 pKa = 10.99YY68 pKa = 10.43VDD70 pKa = 3.42TDD72 pKa = 3.94EE73 pKa = 4.25IDD75 pKa = 3.25RR76 pKa = 11.84RR77 pKa = 11.84VDD79 pKa = 3.05INSASSLKK87 pKa = 9.46HH88 pKa = 5.7TDD90 pKa = 3.25PFLAWFRR97 pKa = 11.84YY98 pKa = 7.77EE99 pKa = 4.5RR100 pKa = 11.84NDD102 pKa = 3.2NGEE105 pKa = 4.35FYY107 pKa = 10.26PARR110 pKa = 11.84DD111 pKa = 3.58MLVSMEE117 pKa = 4.96LLAQCSHH124 pKa = 6.06AMQHH128 pKa = 5.7VLSDD132 pKa = 3.57DD133 pKa = 3.36KK134 pKa = 11.47TVFEE138 pKa = 5.22RR139 pKa = 11.84ILRR142 pKa = 11.84FCSTNCTVNLDD153 pKa = 3.84RR154 pKa = 11.84YY155 pKa = 10.54DD156 pKa = 3.68FLINGPRR163 pKa = 11.84QSIIHH168 pKa = 5.44NTAILSFSLYY178 pKa = 8.91KK179 pKa = 10.04QRR181 pKa = 11.84RR182 pKa = 11.84EE183 pKa = 3.88YY184 pKa = 11.01LKK186 pKa = 10.51EE187 pKa = 3.94FPFPQLQQ194 pKa = 3.1
Molecular weight: 22.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KH10|A0A1L3KH10_9VIRU RdRp catalytic domain-containing protein OS=Wenzhou tombus-like virus 17 OX=1923670 PE=4 SV=1
MM1 pKa = 7.28KK2 pKa = 10.5RR3 pKa = 11.84MGSKK7 pKa = 10.13PPNSDD12 pKa = 3.79GILLSEE18 pKa = 4.62LKK20 pKa = 9.97TFVSEE25 pKa = 4.11WCSKK29 pKa = 10.49NLTPIPPSADD39 pKa = 3.01TSVSAWLDD47 pKa = 3.09KK48 pKa = 10.84CIYY51 pKa = 9.13PKK53 pKa = 9.58WRR55 pKa = 11.84KK56 pKa = 10.03DD57 pKa = 3.37YY58 pKa = 10.73LASLEE63 pKa = 4.36EE64 pKa = 3.96EE65 pKa = 4.58LKK67 pKa = 11.21VKK69 pKa = 10.72LNSTRR74 pKa = 11.84GWNDD78 pKa = 2.3ISKK81 pKa = 10.47VKK83 pKa = 10.72AFTKK87 pKa = 10.04MEE89 pKa = 4.14TYY91 pKa = 10.6PEE93 pKa = 4.19FKK95 pKa = 9.78HH96 pKa = 6.58ARR98 pKa = 11.84GIYY101 pKa = 9.61SRR103 pKa = 11.84SDD105 pKa = 3.15YY106 pKa = 11.06FKK108 pKa = 10.66IKK110 pKa = 9.48TGPIFKK116 pKa = 10.36LIEE119 pKa = 4.18KK120 pKa = 10.0EE121 pKa = 4.37VFKK124 pKa = 10.9HH125 pKa = 6.94DD126 pKa = 3.82YY127 pKa = 9.63FIKK130 pKa = 10.41KK131 pKa = 9.67IPIADD136 pKa = 3.97RR137 pKa = 11.84PSYY140 pKa = 9.69IQEE143 pKa = 3.72HH144 pKa = 6.45VYY146 pKa = 9.86QHH148 pKa = 5.71GAKK151 pKa = 10.28VIATDD156 pKa = 3.74YY157 pKa = 11.43SSFEE161 pKa = 4.03SAFVKK166 pKa = 10.63KK167 pKa = 10.59LMNSCEE173 pKa = 4.02MVMYY177 pKa = 10.38EE178 pKa = 5.36YY179 pKa = 10.1MIQNLPEE186 pKa = 5.01RR187 pKa = 11.84EE188 pKa = 4.06MFLKK192 pKa = 10.48CLNKK196 pKa = 10.03IKK198 pKa = 9.9TKK200 pKa = 10.55QDD202 pKa = 2.74INYY205 pKa = 9.62KK206 pKa = 10.27DD207 pKa = 3.26ININVDD213 pKa = 3.23ATRR216 pKa = 11.84MSGEE220 pKa = 4.05MCTSLGNGFSNLMFTLFLAHH240 pKa = 6.91KK241 pKa = 9.67NGVKK245 pKa = 10.0SLRR248 pKa = 11.84GVVEE252 pKa = 4.08GDD254 pKa = 3.54DD255 pKa = 4.19GLFSFYY261 pKa = 11.57GNLTQEE267 pKa = 4.95HH268 pKa = 6.52YY269 pKa = 10.73SQLGLIIKK277 pKa = 9.87LEE279 pKa = 3.97NVEE282 pKa = 5.15DD283 pKa = 3.72INSASFCGLIFDD295 pKa = 5.03EE296 pKa = 4.66VDD298 pKa = 3.29LSNVTDD304 pKa = 5.07PIEE307 pKa = 4.36VIQNFSWLNPRR318 pKa = 11.84YY319 pKa = 9.5ISSADD324 pKa = 3.57RR325 pKa = 11.84RR326 pKa = 11.84LKK328 pKa = 10.64EE329 pKa = 3.81LLRR332 pKa = 11.84CKK334 pKa = 10.35SLSLAHH340 pKa = 6.25QYY342 pKa = 9.43PGCPIIQSLAHH353 pKa = 5.9YY354 pKa = 9.19GLYY357 pKa = 7.81VTRR360 pKa = 11.84GITIRR365 pKa = 11.84LDD367 pKa = 3.87KK368 pKa = 10.83IHH370 pKa = 6.22MSNWEE375 pKa = 4.01RR376 pKa = 11.84EE377 pKa = 4.12QLILAVNDD385 pKa = 3.28KK386 pKa = 10.21TRR388 pKa = 11.84ILKK391 pKa = 10.32KK392 pKa = 10.46EE393 pKa = 3.56IGNNTRR399 pKa = 11.84LLVEE403 pKa = 4.47RR404 pKa = 11.84KK405 pKa = 9.6FGLTVEE411 pKa = 4.43DD412 pKa = 3.73QIEE415 pKa = 4.12LEE417 pKa = 4.56KK418 pKa = 11.46YY419 pKa = 9.6FDD421 pKa = 3.56SLKK424 pKa = 9.49TVQVLDD430 pKa = 3.91HH431 pKa = 6.69EE432 pKa = 4.79VLTRR436 pKa = 11.84YY437 pKa = 9.84CKK439 pKa = 10.22LQHH442 pKa = 5.85KK443 pKa = 10.21QYY445 pKa = 10.78FNDD448 pKa = 3.47YY449 pKa = 11.04CFVTDD454 pKa = 3.77VNSTNRR460 pKa = 11.84NYY462 pKa = 10.62PIMPRR467 pKa = 11.84RR468 pKa = 11.84DD469 pKa = 3.28FTVNNMSYY477 pKa = 8.3FTEE480 pKa = 4.54HH481 pKa = 6.67KK482 pKa = 10.25SEE484 pKa = 4.62SIRR487 pKa = 11.84KK488 pKa = 9.46SILLRR493 pKa = 4.18
MM1 pKa = 7.28KK2 pKa = 10.5RR3 pKa = 11.84MGSKK7 pKa = 10.13PPNSDD12 pKa = 3.79GILLSEE18 pKa = 4.62LKK20 pKa = 9.97TFVSEE25 pKa = 4.11WCSKK29 pKa = 10.49NLTPIPPSADD39 pKa = 3.01TSVSAWLDD47 pKa = 3.09KK48 pKa = 10.84CIYY51 pKa = 9.13PKK53 pKa = 9.58WRR55 pKa = 11.84KK56 pKa = 10.03DD57 pKa = 3.37YY58 pKa = 10.73LASLEE63 pKa = 4.36EE64 pKa = 3.96EE65 pKa = 4.58LKK67 pKa = 11.21VKK69 pKa = 10.72LNSTRR74 pKa = 11.84GWNDD78 pKa = 2.3ISKK81 pKa = 10.47VKK83 pKa = 10.72AFTKK87 pKa = 10.04MEE89 pKa = 4.14TYY91 pKa = 10.6PEE93 pKa = 4.19FKK95 pKa = 9.78HH96 pKa = 6.58ARR98 pKa = 11.84GIYY101 pKa = 9.61SRR103 pKa = 11.84SDD105 pKa = 3.15YY106 pKa = 11.06FKK108 pKa = 10.66IKK110 pKa = 9.48TGPIFKK116 pKa = 10.36LIEE119 pKa = 4.18KK120 pKa = 10.0EE121 pKa = 4.37VFKK124 pKa = 10.9HH125 pKa = 6.94DD126 pKa = 3.82YY127 pKa = 9.63FIKK130 pKa = 10.41KK131 pKa = 9.67IPIADD136 pKa = 3.97RR137 pKa = 11.84PSYY140 pKa = 9.69IQEE143 pKa = 3.72HH144 pKa = 6.45VYY146 pKa = 9.86QHH148 pKa = 5.71GAKK151 pKa = 10.28VIATDD156 pKa = 3.74YY157 pKa = 11.43SSFEE161 pKa = 4.03SAFVKK166 pKa = 10.63KK167 pKa = 10.59LMNSCEE173 pKa = 4.02MVMYY177 pKa = 10.38EE178 pKa = 5.36YY179 pKa = 10.1MIQNLPEE186 pKa = 5.01RR187 pKa = 11.84EE188 pKa = 4.06MFLKK192 pKa = 10.48CLNKK196 pKa = 10.03IKK198 pKa = 9.9TKK200 pKa = 10.55QDD202 pKa = 2.74INYY205 pKa = 9.62KK206 pKa = 10.27DD207 pKa = 3.26ININVDD213 pKa = 3.23ATRR216 pKa = 11.84MSGEE220 pKa = 4.05MCTSLGNGFSNLMFTLFLAHH240 pKa = 6.91KK241 pKa = 9.67NGVKK245 pKa = 10.0SLRR248 pKa = 11.84GVVEE252 pKa = 4.08GDD254 pKa = 3.54DD255 pKa = 4.19GLFSFYY261 pKa = 11.57GNLTQEE267 pKa = 4.95HH268 pKa = 6.52YY269 pKa = 10.73SQLGLIIKK277 pKa = 9.87LEE279 pKa = 3.97NVEE282 pKa = 5.15DD283 pKa = 3.72INSASFCGLIFDD295 pKa = 5.03EE296 pKa = 4.66VDD298 pKa = 3.29LSNVTDD304 pKa = 5.07PIEE307 pKa = 4.36VIQNFSWLNPRR318 pKa = 11.84YY319 pKa = 9.5ISSADD324 pKa = 3.57RR325 pKa = 11.84RR326 pKa = 11.84LKK328 pKa = 10.64EE329 pKa = 3.81LLRR332 pKa = 11.84CKK334 pKa = 10.35SLSLAHH340 pKa = 6.25QYY342 pKa = 9.43PGCPIIQSLAHH353 pKa = 5.9YY354 pKa = 9.19GLYY357 pKa = 7.81VTRR360 pKa = 11.84GITIRR365 pKa = 11.84LDD367 pKa = 3.87KK368 pKa = 10.83IHH370 pKa = 6.22MSNWEE375 pKa = 4.01RR376 pKa = 11.84EE377 pKa = 4.12QLILAVNDD385 pKa = 3.28KK386 pKa = 10.21TRR388 pKa = 11.84ILKK391 pKa = 10.32KK392 pKa = 10.46EE393 pKa = 3.56IGNNTRR399 pKa = 11.84LLVEE403 pKa = 4.47RR404 pKa = 11.84KK405 pKa = 9.6FGLTVEE411 pKa = 4.43DD412 pKa = 3.73QIEE415 pKa = 4.12LEE417 pKa = 4.56KK418 pKa = 11.46YY419 pKa = 9.6FDD421 pKa = 3.56SLKK424 pKa = 9.49TVQVLDD430 pKa = 3.91HH431 pKa = 6.69EE432 pKa = 4.79VLTRR436 pKa = 11.84YY437 pKa = 9.84CKK439 pKa = 10.22LQHH442 pKa = 5.85KK443 pKa = 10.21QYY445 pKa = 10.78FNDD448 pKa = 3.47YY449 pKa = 11.04CFVTDD454 pKa = 3.77VNSTNRR460 pKa = 11.84NYY462 pKa = 10.62PIMPRR467 pKa = 11.84RR468 pKa = 11.84DD469 pKa = 3.28FTVNNMSYY477 pKa = 8.3FTEE480 pKa = 4.54HH481 pKa = 6.67KK482 pKa = 10.25SEE484 pKa = 4.62SIRR487 pKa = 11.84KK488 pKa = 9.46SILLRR493 pKa = 4.18
Molecular weight: 57.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
687 |
194 |
493 |
343.5 |
40.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.639 ± 0.554 | 2.183 ± 0.218 |
5.968 ± 0.692 | 5.822 ± 1.227 |
5.531 ± 0.934 | 4.076 ± 0.259 |
2.62 ± 0.262 | 7.278 ± 0.32 |
7.278 ± 2.319 | 9.461 ± 0.387 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.057 ± 0.306 | 5.531 ± 0.494 |
3.639 ± 0.269 | 3.493 ± 0.921 |
5.531 ± 0.648 | 7.569 ± 0.767 |
5.24 ± 0.238 | 5.531 ± 0.077 |
1.164 ± 0.074 | 5.386 ± 0.729 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
