
Human feces-associated smacovirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus; unclassified Porprismacovirus
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
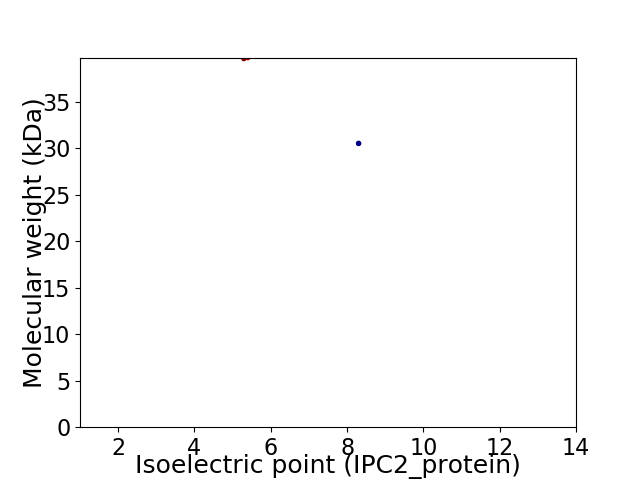
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I9WIN8|A0A1I9WIN8_9VIRU Replication associated protein OS=Human feces-associated smacovirus OX=1914415 PE=4 SV=1
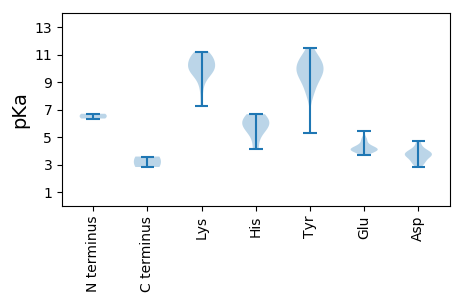
MM1 pKa = 6.69ATNYY5 pKa = 10.66ASASYY10 pKa = 10.76QEE12 pKa = 4.29ILDD15 pKa = 3.88VNTVKK20 pKa = 11.08GNVTVIGIHH29 pKa = 5.93TPTGSTPVRR38 pKa = 11.84RR39 pKa = 11.84LSGFFAQFRR48 pKa = 11.84KK49 pKa = 9.57FRR51 pKa = 11.84YY52 pKa = 8.89KK53 pKa = 9.37GCSVQVVPAATLPADD68 pKa = 3.8PLQVGFEE75 pKa = 4.04AGEE78 pKa = 3.97NQIDD82 pKa = 3.95MRR84 pKa = 11.84DD85 pKa = 3.58MLNPILFHH93 pKa = 5.87GTHH96 pKa = 6.41GEE98 pKa = 4.12SLQVAWNNIFVNPNRR113 pKa = 11.84FDD115 pKa = 3.55GSASGNSSYY124 pKa = 9.56KK125 pKa = 9.97TGAGFDD131 pKa = 3.71ASDD134 pKa = 3.6LTFADD139 pKa = 3.7STNNPVEE146 pKa = 4.12TQYY149 pKa = 11.45YY150 pKa = 8.61QAITDD155 pKa = 3.78RR156 pKa = 11.84TWRR159 pKa = 11.84KK160 pKa = 9.59FGIQSTFKK168 pKa = 10.76LPFMSPRR175 pKa = 11.84VWKK178 pKa = 10.52ASTIYY183 pKa = 10.26PILPNGYY190 pKa = 7.94QQNGSMNTGDD200 pKa = 3.58TPLPEE205 pKa = 4.98GFLPPGTYY213 pKa = 9.77FDD215 pKa = 4.72NYY217 pKa = 10.42DD218 pKa = 3.45GRR220 pKa = 11.84GLTMPIDD227 pKa = 4.34GQGHH231 pKa = 6.43ILDD234 pKa = 4.16QKK236 pKa = 9.63WMSSGTTPLGWLPTVTMPRR255 pKa = 11.84VDD257 pKa = 3.95PNNATSVTLAPRR269 pKa = 11.84TLLPKK274 pKa = 9.91IFMGILVLPPSYY286 pKa = 10.47LQEE289 pKa = 3.97LYY291 pKa = 10.53FRR293 pKa = 11.84VIVTHH298 pKa = 5.85YY299 pKa = 10.86YY300 pKa = 9.28EE301 pKa = 4.74FKK303 pKa = 11.15GFTASLAPASEE314 pKa = 4.23MPDD317 pKa = 3.11VKK319 pKa = 10.23GTYY322 pKa = 9.78EE323 pKa = 3.93NAVKK327 pKa = 8.76QTAATSNVMALDD339 pKa = 3.77EE340 pKa = 4.83VEE342 pKa = 5.46DD343 pKa = 3.97STLEE347 pKa = 4.06SPTANVVRR355 pKa = 11.84ISDD358 pKa = 3.85GVFF361 pKa = 2.82
MM1 pKa = 6.69ATNYY5 pKa = 10.66ASASYY10 pKa = 10.76QEE12 pKa = 4.29ILDD15 pKa = 3.88VNTVKK20 pKa = 11.08GNVTVIGIHH29 pKa = 5.93TPTGSTPVRR38 pKa = 11.84RR39 pKa = 11.84LSGFFAQFRR48 pKa = 11.84KK49 pKa = 9.57FRR51 pKa = 11.84YY52 pKa = 8.89KK53 pKa = 9.37GCSVQVVPAATLPADD68 pKa = 3.8PLQVGFEE75 pKa = 4.04AGEE78 pKa = 3.97NQIDD82 pKa = 3.95MRR84 pKa = 11.84DD85 pKa = 3.58MLNPILFHH93 pKa = 5.87GTHH96 pKa = 6.41GEE98 pKa = 4.12SLQVAWNNIFVNPNRR113 pKa = 11.84FDD115 pKa = 3.55GSASGNSSYY124 pKa = 9.56KK125 pKa = 9.97TGAGFDD131 pKa = 3.71ASDD134 pKa = 3.6LTFADD139 pKa = 3.7STNNPVEE146 pKa = 4.12TQYY149 pKa = 11.45YY150 pKa = 8.61QAITDD155 pKa = 3.78RR156 pKa = 11.84TWRR159 pKa = 11.84KK160 pKa = 9.59FGIQSTFKK168 pKa = 10.76LPFMSPRR175 pKa = 11.84VWKK178 pKa = 10.52ASTIYY183 pKa = 10.26PILPNGYY190 pKa = 7.94QQNGSMNTGDD200 pKa = 3.58TPLPEE205 pKa = 4.98GFLPPGTYY213 pKa = 9.77FDD215 pKa = 4.72NYY217 pKa = 10.42DD218 pKa = 3.45GRR220 pKa = 11.84GLTMPIDD227 pKa = 4.34GQGHH231 pKa = 6.43ILDD234 pKa = 4.16QKK236 pKa = 9.63WMSSGTTPLGWLPTVTMPRR255 pKa = 11.84VDD257 pKa = 3.95PNNATSVTLAPRR269 pKa = 11.84TLLPKK274 pKa = 9.91IFMGILVLPPSYY286 pKa = 10.47LQEE289 pKa = 3.97LYY291 pKa = 10.53FRR293 pKa = 11.84VIVTHH298 pKa = 5.85YY299 pKa = 10.86YY300 pKa = 9.28EE301 pKa = 4.74FKK303 pKa = 11.15GFTASLAPASEE314 pKa = 4.23MPDD317 pKa = 3.11VKK319 pKa = 10.23GTYY322 pKa = 9.78EE323 pKa = 3.93NAVKK327 pKa = 8.76QTAATSNVMALDD339 pKa = 3.77EE340 pKa = 4.83VEE342 pKa = 5.46DD343 pKa = 3.97STLEE347 pKa = 4.06SPTANVVRR355 pKa = 11.84ISDD358 pKa = 3.85GVFF361 pKa = 2.82
Molecular weight: 39.66 kDa
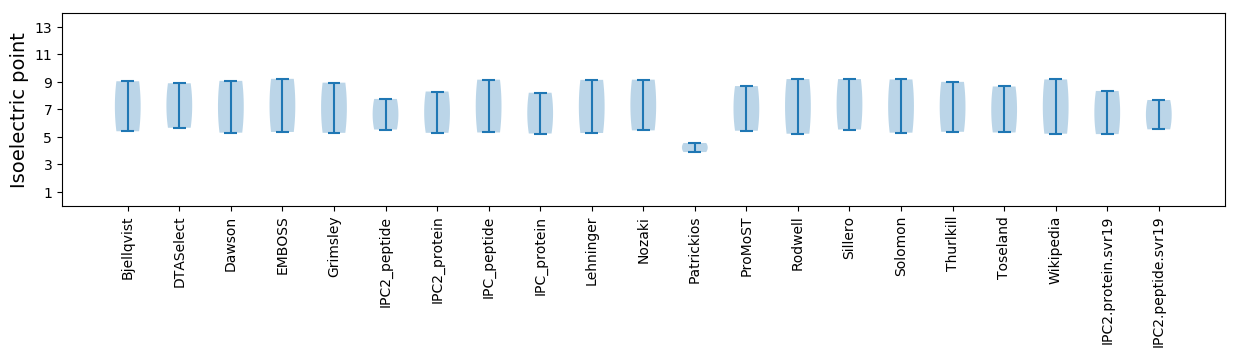
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I9WIN8|A0A1I9WIN8_9VIRU Replication associated protein OS=Human feces-associated smacovirus OX=1914415 PE=4 SV=1
MM1 pKa = 6.35VQAWMITGPRR11 pKa = 11.84SEE13 pKa = 4.42SIEE16 pKa = 3.92VEE18 pKa = 4.5EE19 pKa = 4.17IGGWQIEE26 pKa = 3.8IDD28 pKa = 3.35RR29 pKa = 11.84RR30 pKa = 11.84ISKK33 pKa = 9.72KK34 pKa = 10.61AIMIMLSKK42 pKa = 10.62NDD44 pKa = 3.71CKK46 pKa = 10.78KK47 pKa = 8.03WTIGQEE53 pKa = 4.03VGKK56 pKa = 10.15NGYY59 pKa = 8.23KK60 pKa = 9.35HH61 pKa = 4.12WQIRR65 pKa = 11.84VEE67 pKa = 4.1SSNTNFFKK75 pKa = 10.03WCKK78 pKa = 7.25QHH80 pKa = 6.66IPSAHH85 pKa = 5.08VEE87 pKa = 4.06KK88 pKa = 10.93AEE90 pKa = 4.13KK91 pKa = 10.64GVDD94 pKa = 3.16EE95 pKa = 4.52SRR97 pKa = 11.84YY98 pKa = 5.3EE99 pKa = 4.21TKK101 pKa = 10.38EE102 pKa = 3.86GQYY105 pKa = 8.52TQYY108 pKa = 10.01TDD110 pKa = 2.84RR111 pKa = 11.84VEE113 pKa = 3.89ILKK116 pKa = 9.93QRR118 pKa = 11.84FGKK121 pKa = 8.87MRR123 pKa = 11.84PNQIRR128 pKa = 11.84ALEE131 pKa = 4.11ALEE134 pKa = 4.1ATNDD138 pKa = 3.59RR139 pKa = 11.84EE140 pKa = 4.21VLVWYY145 pKa = 9.95DD146 pKa = 3.17EE147 pKa = 4.28SGNVGKK153 pKa = 10.44SWLTGALWEE162 pKa = 4.84RR163 pKa = 11.84GLAYY167 pKa = 9.61YY168 pKa = 9.54VPPTVDD174 pKa = 2.8SVKK177 pKa = 11.22AMVQWVASCYY187 pKa = 9.73QSGGWRR193 pKa = 11.84PYY195 pKa = 10.33IIIDD199 pKa = 4.57IPRR202 pKa = 11.84SWKK205 pKa = 9.69WSEE208 pKa = 3.68QLYY211 pKa = 10.73AAIEE215 pKa = 4.33SIKK218 pKa = 10.97DD219 pKa = 3.11GLVYY223 pKa = 9.39DD224 pKa = 4.17TRR226 pKa = 11.84YY227 pKa = 9.55HH228 pKa = 5.6AQCINIRR235 pKa = 11.84GVKK238 pKa = 10.09VLVLTNTKK246 pKa = 10.11PKK248 pKa = 10.19LDD250 pKa = 3.95KK251 pKa = 11.04LSADD255 pKa = 3.24RR256 pKa = 11.84WRR258 pKa = 11.84ICVFF262 pKa = 3.58
MM1 pKa = 6.35VQAWMITGPRR11 pKa = 11.84SEE13 pKa = 4.42SIEE16 pKa = 3.92VEE18 pKa = 4.5EE19 pKa = 4.17IGGWQIEE26 pKa = 3.8IDD28 pKa = 3.35RR29 pKa = 11.84RR30 pKa = 11.84ISKK33 pKa = 9.72KK34 pKa = 10.61AIMIMLSKK42 pKa = 10.62NDD44 pKa = 3.71CKK46 pKa = 10.78KK47 pKa = 8.03WTIGQEE53 pKa = 4.03VGKK56 pKa = 10.15NGYY59 pKa = 8.23KK60 pKa = 9.35HH61 pKa = 4.12WQIRR65 pKa = 11.84VEE67 pKa = 4.1SSNTNFFKK75 pKa = 10.03WCKK78 pKa = 7.25QHH80 pKa = 6.66IPSAHH85 pKa = 5.08VEE87 pKa = 4.06KK88 pKa = 10.93AEE90 pKa = 4.13KK91 pKa = 10.64GVDD94 pKa = 3.16EE95 pKa = 4.52SRR97 pKa = 11.84YY98 pKa = 5.3EE99 pKa = 4.21TKK101 pKa = 10.38EE102 pKa = 3.86GQYY105 pKa = 8.52TQYY108 pKa = 10.01TDD110 pKa = 2.84RR111 pKa = 11.84VEE113 pKa = 3.89ILKK116 pKa = 9.93QRR118 pKa = 11.84FGKK121 pKa = 8.87MRR123 pKa = 11.84PNQIRR128 pKa = 11.84ALEE131 pKa = 4.11ALEE134 pKa = 4.1ATNDD138 pKa = 3.59RR139 pKa = 11.84EE140 pKa = 4.21VLVWYY145 pKa = 9.95DD146 pKa = 3.17EE147 pKa = 4.28SGNVGKK153 pKa = 10.44SWLTGALWEE162 pKa = 4.84RR163 pKa = 11.84GLAYY167 pKa = 9.61YY168 pKa = 9.54VPPTVDD174 pKa = 2.8SVKK177 pKa = 11.22AMVQWVASCYY187 pKa = 9.73QSGGWRR193 pKa = 11.84PYY195 pKa = 10.33IIIDD199 pKa = 4.57IPRR202 pKa = 11.84SWKK205 pKa = 9.69WSEE208 pKa = 3.68QLYY211 pKa = 10.73AAIEE215 pKa = 4.33SIKK218 pKa = 10.97DD219 pKa = 3.11GLVYY223 pKa = 9.39DD224 pKa = 4.17TRR226 pKa = 11.84YY227 pKa = 9.55HH228 pKa = 5.6AQCINIRR235 pKa = 11.84GVKK238 pKa = 10.09VLVLTNTKK246 pKa = 10.11PKK248 pKa = 10.19LDD250 pKa = 3.95KK251 pKa = 11.04LSADD255 pKa = 3.24RR256 pKa = 11.84WRR258 pKa = 11.84ICVFF262 pKa = 3.58
Molecular weight: 30.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
623 |
262 |
361 |
311.5 |
35.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.421 ± 0.433 | 0.963 ± 0.589 |
4.976 ± 0.247 | 5.297 ± 1.456 |
3.852 ± 1.449 | 7.384 ± 0.558 |
1.445 ± 0.051 | 6.1 ± 1.432 |
5.457 ± 1.832 | 6.421 ± 0.671 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.729 ± 0.273 | 4.815 ± 0.86 |
5.778 ± 1.698 | 4.494 ± 0.291 |
4.976 ± 0.943 | 6.902 ± 0.258 |
7.384 ± 1.747 | 7.384 ± 0.156 |
2.889 ± 1.292 | 4.334 ± 0.153 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
