
Pyrrhula pyrrhula polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Gammapolyomavirus
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
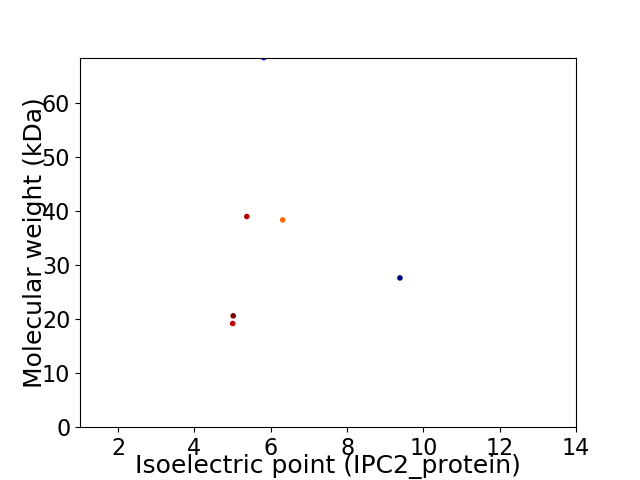
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
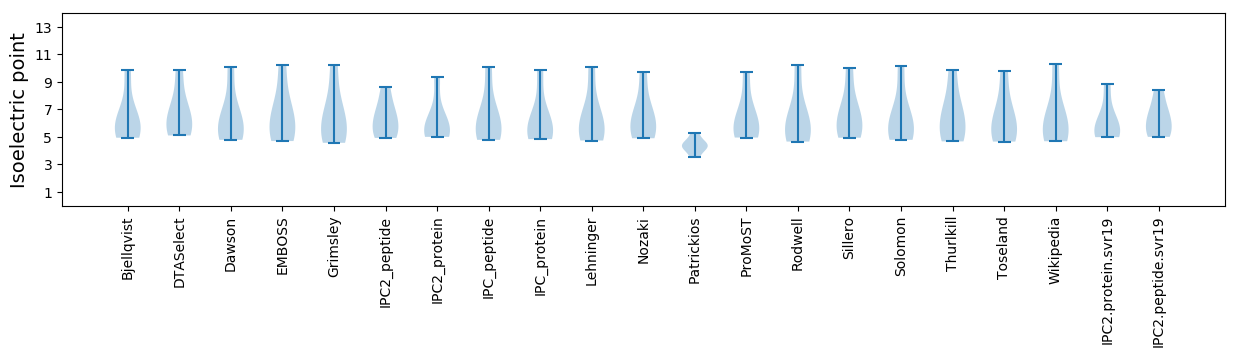
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2I6RKB0|A0A2I6RKB0_9POLY Capsid protein VP1 OS=Pyrrhula pyrrhula polyomavirus 1 OX=1891751 PE=3 SV=1
MM1 pKa = 7.27STLQKK6 pKa = 10.89LIDD9 pKa = 4.37LLGLPPSATEE19 pKa = 3.5ADD21 pKa = 3.51VRR23 pKa = 11.84SAYY26 pKa = 9.89RR27 pKa = 11.84KK28 pKa = 9.5KK29 pKa = 10.66ALEE32 pKa = 3.77FHH34 pKa = 7.12PDD36 pKa = 3.04KK37 pKa = 11.5GGDD40 pKa = 3.6PEE42 pKa = 5.25RR43 pKa = 11.84MKK45 pKa = 10.73EE46 pKa = 3.98LNRR49 pKa = 11.84LMDD52 pKa = 4.19EE53 pKa = 4.43FRR55 pKa = 11.84HH56 pKa = 5.32SQSLFCDD63 pKa = 3.38EE64 pKa = 4.89TLDD67 pKa = 4.59SDD69 pKa = 5.11SDD71 pKa = 4.4DD72 pKa = 4.11GDD74 pKa = 3.9SPGPSQRR81 pKa = 11.84TSTPEE86 pKa = 3.81PGTGKK91 pKa = 10.39DD92 pKa = 3.25SGHH95 pKa = 5.77GTFEE99 pKa = 4.29PEE101 pKa = 3.84VNSWHH106 pKa = 7.14AIDD109 pKa = 4.97YY110 pKa = 10.56DD111 pKa = 3.64RR112 pKa = 11.84AYY114 pKa = 10.53CRR116 pKa = 11.84LMEE119 pKa = 4.96LKK121 pKa = 9.57WCLEE125 pKa = 4.02SFFTSTEE132 pKa = 3.57RR133 pKa = 11.84RR134 pKa = 11.84KK135 pKa = 10.21QGLTPEE141 pKa = 3.97YY142 pKa = 10.67LEE144 pKa = 4.28LKK146 pKa = 10.41RR147 pKa = 11.84RR148 pKa = 11.84FQAVPWRR155 pKa = 11.84VFDD158 pKa = 4.01NVFNMDD164 pKa = 4.08YY165 pKa = 10.73II166 pKa = 4.95
MM1 pKa = 7.27STLQKK6 pKa = 10.89LIDD9 pKa = 4.37LLGLPPSATEE19 pKa = 3.5ADD21 pKa = 3.51VRR23 pKa = 11.84SAYY26 pKa = 9.89RR27 pKa = 11.84KK28 pKa = 9.5KK29 pKa = 10.66ALEE32 pKa = 3.77FHH34 pKa = 7.12PDD36 pKa = 3.04KK37 pKa = 11.5GGDD40 pKa = 3.6PEE42 pKa = 5.25RR43 pKa = 11.84MKK45 pKa = 10.73EE46 pKa = 3.98LNRR49 pKa = 11.84LMDD52 pKa = 4.19EE53 pKa = 4.43FRR55 pKa = 11.84HH56 pKa = 5.32SQSLFCDD63 pKa = 3.38EE64 pKa = 4.89TLDD67 pKa = 4.59SDD69 pKa = 5.11SDD71 pKa = 4.4DD72 pKa = 4.11GDD74 pKa = 3.9SPGPSQRR81 pKa = 11.84TSTPEE86 pKa = 3.81PGTGKK91 pKa = 10.39DD92 pKa = 3.25SGHH95 pKa = 5.77GTFEE99 pKa = 4.29PEE101 pKa = 3.84VNSWHH106 pKa = 7.14AIDD109 pKa = 4.97YY110 pKa = 10.56DD111 pKa = 3.64RR112 pKa = 11.84AYY114 pKa = 10.53CRR116 pKa = 11.84LMEE119 pKa = 4.96LKK121 pKa = 9.57WCLEE125 pKa = 4.02SFFTSTEE132 pKa = 3.57RR133 pKa = 11.84RR134 pKa = 11.84KK135 pKa = 10.21QGLTPEE141 pKa = 3.97YY142 pKa = 10.67LEE144 pKa = 4.28LKK146 pKa = 10.41RR147 pKa = 11.84RR148 pKa = 11.84FQAVPWRR155 pKa = 11.84VFDD158 pKa = 4.01NVFNMDD164 pKa = 4.08YY165 pKa = 10.73II166 pKa = 4.95
Molecular weight: 19.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q20HX9|Q20HX9_9POLY Minor capsid protein OS=Pyrrhula pyrrhula polyomavirus 1 OX=1891751 PE=3 SV=1
MM1 pKa = 7.56ALAIWRR7 pKa = 11.84EE8 pKa = 3.97QIDD11 pKa = 3.93YY12 pKa = 10.33LFPGINWLATNIHH25 pKa = 6.16YY26 pKa = 10.56LDD28 pKa = 4.24PLHH31 pKa = 6.95WARR34 pKa = 11.84SLFNQVGRR42 pKa = 11.84ALWNQIDD49 pKa = 3.6QRR51 pKa = 11.84VRR53 pKa = 11.84DD54 pKa = 3.97GLIGGAAQAAGAAGAEE70 pKa = 4.22VAIRR74 pKa = 11.84QSRR77 pKa = 11.84ALYY80 pKa = 10.22DD81 pKa = 3.4VLARR85 pKa = 11.84AMEE88 pKa = 4.06TARR91 pKa = 11.84WTIHH95 pKa = 5.85SSYY98 pKa = 10.67TSAEE102 pKa = 3.61EE103 pKa = 4.12TYY105 pKa = 10.74RR106 pKa = 11.84NLRR109 pKa = 11.84EE110 pKa = 4.19YY111 pKa = 10.53YY112 pKa = 10.41AQLPVDD118 pKa = 4.55AGRR121 pKa = 11.84PAYY124 pKa = 9.88RR125 pKa = 11.84RR126 pKa = 11.84RR127 pKa = 11.84LLGLTEE133 pKa = 3.92QSSWDD138 pKa = 3.51HH139 pKa = 5.8GRR141 pKa = 11.84GPTTSTTEE149 pKa = 3.97PKK151 pKa = 10.62AKK153 pKa = 10.33AIEE156 pKa = 4.24EE157 pKa = 4.12ASKK160 pKa = 10.58EE161 pKa = 4.05PPEE164 pKa = 4.63SGEE167 pKa = 4.13HH168 pKa = 5.01VQDD171 pKa = 3.8YY172 pKa = 8.43PPPGGAHH179 pKa = 5.41QRR181 pKa = 11.84HH182 pKa = 5.67APDD185 pKa = 3.0WLLPLLLGLYY195 pKa = 10.7GDD197 pKa = 5.19LTPEE201 pKa = 3.55WRR203 pKa = 11.84SQLKK207 pKa = 9.25QLGYY211 pKa = 10.8GSQKK215 pKa = 10.34RR216 pKa = 11.84KK217 pKa = 9.48RR218 pKa = 11.84QLSPTPASPQADD230 pKa = 3.43SKK232 pKa = 11.05RR233 pKa = 11.84RR234 pKa = 11.84NRR236 pKa = 11.84STRR239 pKa = 11.84RR240 pKa = 11.84KK241 pKa = 9.56NRR243 pKa = 11.84PP244 pKa = 2.95
MM1 pKa = 7.56ALAIWRR7 pKa = 11.84EE8 pKa = 3.97QIDD11 pKa = 3.93YY12 pKa = 10.33LFPGINWLATNIHH25 pKa = 6.16YY26 pKa = 10.56LDD28 pKa = 4.24PLHH31 pKa = 6.95WARR34 pKa = 11.84SLFNQVGRR42 pKa = 11.84ALWNQIDD49 pKa = 3.6QRR51 pKa = 11.84VRR53 pKa = 11.84DD54 pKa = 3.97GLIGGAAQAAGAAGAEE70 pKa = 4.22VAIRR74 pKa = 11.84QSRR77 pKa = 11.84ALYY80 pKa = 10.22DD81 pKa = 3.4VLARR85 pKa = 11.84AMEE88 pKa = 4.06TARR91 pKa = 11.84WTIHH95 pKa = 5.85SSYY98 pKa = 10.67TSAEE102 pKa = 3.61EE103 pKa = 4.12TYY105 pKa = 10.74RR106 pKa = 11.84NLRR109 pKa = 11.84EE110 pKa = 4.19YY111 pKa = 10.53YY112 pKa = 10.41AQLPVDD118 pKa = 4.55AGRR121 pKa = 11.84PAYY124 pKa = 9.88RR125 pKa = 11.84RR126 pKa = 11.84RR127 pKa = 11.84LLGLTEE133 pKa = 3.92QSSWDD138 pKa = 3.51HH139 pKa = 5.8GRR141 pKa = 11.84GPTTSTTEE149 pKa = 3.97PKK151 pKa = 10.62AKK153 pKa = 10.33AIEE156 pKa = 4.24EE157 pKa = 4.12ASKK160 pKa = 10.58EE161 pKa = 4.05PPEE164 pKa = 4.63SGEE167 pKa = 4.13HH168 pKa = 5.01VQDD171 pKa = 3.8YY172 pKa = 8.43PPPGGAHH179 pKa = 5.41QRR181 pKa = 11.84HH182 pKa = 5.67APDD185 pKa = 3.0WLLPLLLGLYY195 pKa = 10.7GDD197 pKa = 5.19LTPEE201 pKa = 3.55WRR203 pKa = 11.84SQLKK207 pKa = 9.25QLGYY211 pKa = 10.8GSQKK215 pKa = 10.34RR216 pKa = 11.84KK217 pKa = 9.48RR218 pKa = 11.84QLSPTPASPQADD230 pKa = 3.43SKK232 pKa = 11.05RR233 pKa = 11.84RR234 pKa = 11.84NRR236 pKa = 11.84STRR239 pKa = 11.84RR240 pKa = 11.84KK241 pKa = 9.56NRR243 pKa = 11.84PP244 pKa = 2.95
Molecular weight: 27.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1939 |
166 |
612 |
323.2 |
35.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.664 ± 1.445 | 1.392 ± 0.453 |
5.725 ± 0.662 | 5.931 ± 0.462 |
2.527 ± 0.507 | 8.045 ± 1.038 |
2.269 ± 0.384 | 3.92 ± 0.51 |
4.126 ± 0.587 | 10.057 ± 0.714 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.908 ± 0.333 | 3.559 ± 0.432 |
7.684 ± 1.336 | 4.899 ± 0.326 |
6.601 ± 0.865 | 6.962 ± 0.856 |
6.756 ± 0.494 | 4.642 ± 0.713 |
1.289 ± 0.417 | 3.043 ± 0.463 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
