
Motilimonas pumilua
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadales genera incertae sedis; Motilimonas
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
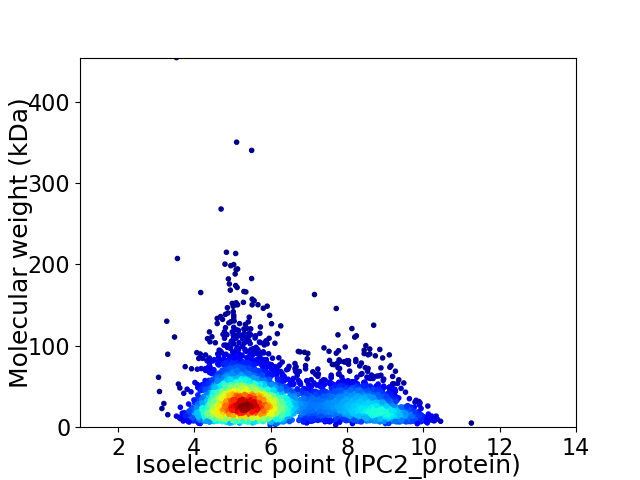
Virtual 2D-PAGE plot for 4087 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
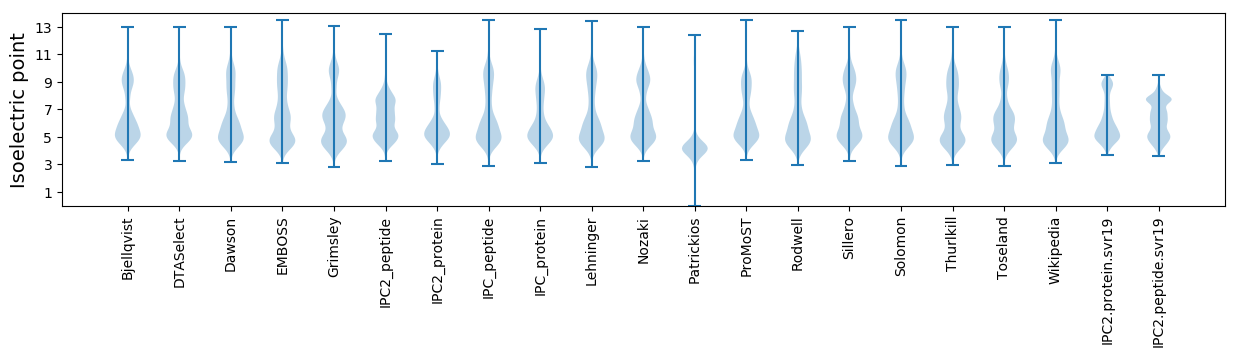
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A418YH71|A0A418YH71_9GAMM Uncharacterized protein OS=Motilimonas pumilua OX=2303987 GN=D1Z90_05670 PE=4 SV=1
MM1 pKa = 8.12LMFKK5 pKa = 10.42KK6 pKa = 10.63APLYY10 pKa = 10.31LALSAAIALVGCGGSGSSSDD30 pKa = 4.42DD31 pKa = 3.28SDD33 pKa = 3.91GGSTSVEE40 pKa = 3.86PTLAPVVTPSPAPVTTATEE59 pKa = 3.94PRR61 pKa = 11.84SVKK64 pKa = 10.5GVINAPGLTDD74 pKa = 3.25SSVFKK79 pKa = 10.8ISLVSLNAAGEE90 pKa = 4.08LVQQFEE96 pKa = 4.48EE97 pKa = 4.28QLQQTMGQVLFDD109 pKa = 3.97SAIQLASDD117 pKa = 4.07GGSLVVTASSDD128 pKa = 3.4GYY130 pKa = 11.35ASYY133 pKa = 11.57SKK135 pKa = 9.83TFDD138 pKa = 3.1FDD140 pKa = 3.49AASDD144 pKa = 4.12FNSEE148 pKa = 4.14LQVNLQQVNQVVVQKK163 pKa = 10.82GNTIARR169 pKa = 11.84SGLRR173 pKa = 11.84QDD175 pKa = 3.62TFSFAVHH182 pKa = 6.55KK183 pKa = 9.49MPSGRR188 pKa = 11.84SIVVDD193 pKa = 3.6GNGPVPRR200 pKa = 11.84NSLMEE205 pKa = 4.84EE206 pKa = 3.52IRR208 pKa = 11.84IDD210 pKa = 3.44IPADD214 pKa = 3.32SLPASTSALSGRR226 pKa = 11.84IANYY230 pKa = 10.5DD231 pKa = 3.26PTDD234 pKa = 3.82AEE236 pKa = 4.11EE237 pKa = 5.13SEE239 pKa = 4.74AFPGAYY245 pKa = 8.96RR246 pKa = 11.84DD247 pKa = 3.61SDD249 pKa = 3.8GNTLVSVAFSFAEE262 pKa = 4.03IRR264 pKa = 11.84DD265 pKa = 3.58QDD267 pKa = 3.77GQTLSGAIARR277 pKa = 11.84AAEE280 pKa = 4.0QGEE283 pKa = 4.49LVARR287 pKa = 11.84NAAPTMITRR296 pKa = 11.84TIPAGSCASLEE307 pKa = 4.0QLQDD311 pKa = 3.5ANSDD315 pKa = 3.34KK316 pKa = 11.41AGFQIPVYY324 pKa = 9.49TYY326 pKa = 11.03NPNAGDD332 pKa = 3.87WDD334 pKa = 4.13LLGYY338 pKa = 11.06GDD340 pKa = 4.7VFNAADD346 pKa = 4.23DD347 pKa = 4.74SIVPDD352 pKa = 3.51NQQVFDD358 pKa = 4.68CPADD362 pKa = 3.42NYY364 pKa = 9.86YY365 pKa = 11.12LEE367 pKa = 4.77VVVSNDD373 pKa = 3.81DD374 pKa = 4.56FLRR377 pKa = 11.84DD378 pKa = 3.47WWNLDD383 pKa = 3.31YY384 pKa = 10.87PLLFDD389 pKa = 4.89TPVEE393 pKa = 3.84LCANVKK399 pKa = 9.91LLNPSGNPVAGSYY412 pKa = 10.77VFLSDD417 pKa = 3.43PSGNRR422 pKa = 11.84SFSDD426 pKa = 3.85TYY428 pKa = 11.61AVTDD432 pKa = 4.12GQGSVALSTTLLNPDD447 pKa = 3.03NTQRR451 pKa = 11.84TASLRR456 pKa = 11.84HH457 pKa = 5.86WGYY460 pKa = 11.1QSGTYY465 pKa = 9.34LASDD469 pKa = 3.47IQLSEE474 pKa = 4.09QEE476 pKa = 4.28SCGEE480 pKa = 4.02VQVITVQKK488 pKa = 10.57PDD490 pKa = 3.29MCSVSGTVTRR500 pKa = 11.84AAGGSTDD507 pKa = 4.2EE508 pKa = 4.18NSGWLYY514 pKa = 11.01AFPTNTEE521 pKa = 3.72GYY523 pKa = 8.67MNFGFTQAQANGQYY537 pKa = 11.32SMDD540 pKa = 3.82VEE542 pKa = 4.5CDD544 pKa = 2.65IDD546 pKa = 3.79YY547 pKa = 11.18QFYY550 pKa = 9.56TFSDD554 pKa = 5.17FYY556 pKa = 11.33QGINAEE562 pKa = 4.13QEE564 pKa = 4.16AQAKK568 pKa = 6.6TFNVNGTVSGFEE580 pKa = 4.07TSDD583 pKa = 4.02DD584 pKa = 3.51GDD586 pKa = 3.68KK587 pKa = 11.28AILQDD592 pKa = 5.26AEE594 pKa = 4.24IQNLAPLAILTGHH607 pKa = 7.38IMPQGQSMSFLLMDD621 pKa = 4.55FDD623 pKa = 4.71FDD625 pKa = 4.44YY626 pKa = 10.81PVSYY630 pKa = 10.58RR631 pKa = 11.84LEE633 pKa = 4.04FTDD636 pKa = 3.27MNGAVVASVEE646 pKa = 4.04GTAANEE652 pKa = 3.5IDD654 pKa = 4.34YY655 pKa = 10.94DD656 pKa = 4.27SEE658 pKa = 4.68DD659 pKa = 3.32MGAAPFTLPTDD670 pKa = 4.25LPLGSYY676 pKa = 9.69SVSGILTDD684 pKa = 3.35ATGNQGSASGIIGVTEE700 pKa = 3.81
MM1 pKa = 8.12LMFKK5 pKa = 10.42KK6 pKa = 10.63APLYY10 pKa = 10.31LALSAAIALVGCGGSGSSSDD30 pKa = 4.42DD31 pKa = 3.28SDD33 pKa = 3.91GGSTSVEE40 pKa = 3.86PTLAPVVTPSPAPVTTATEE59 pKa = 3.94PRR61 pKa = 11.84SVKK64 pKa = 10.5GVINAPGLTDD74 pKa = 3.25SSVFKK79 pKa = 10.8ISLVSLNAAGEE90 pKa = 4.08LVQQFEE96 pKa = 4.48EE97 pKa = 4.28QLQQTMGQVLFDD109 pKa = 3.97SAIQLASDD117 pKa = 4.07GGSLVVTASSDD128 pKa = 3.4GYY130 pKa = 11.35ASYY133 pKa = 11.57SKK135 pKa = 9.83TFDD138 pKa = 3.1FDD140 pKa = 3.49AASDD144 pKa = 4.12FNSEE148 pKa = 4.14LQVNLQQVNQVVVQKK163 pKa = 10.82GNTIARR169 pKa = 11.84SGLRR173 pKa = 11.84QDD175 pKa = 3.62TFSFAVHH182 pKa = 6.55KK183 pKa = 9.49MPSGRR188 pKa = 11.84SIVVDD193 pKa = 3.6GNGPVPRR200 pKa = 11.84NSLMEE205 pKa = 4.84EE206 pKa = 3.52IRR208 pKa = 11.84IDD210 pKa = 3.44IPADD214 pKa = 3.32SLPASTSALSGRR226 pKa = 11.84IANYY230 pKa = 10.5DD231 pKa = 3.26PTDD234 pKa = 3.82AEE236 pKa = 4.11EE237 pKa = 5.13SEE239 pKa = 4.74AFPGAYY245 pKa = 8.96RR246 pKa = 11.84DD247 pKa = 3.61SDD249 pKa = 3.8GNTLVSVAFSFAEE262 pKa = 4.03IRR264 pKa = 11.84DD265 pKa = 3.58QDD267 pKa = 3.77GQTLSGAIARR277 pKa = 11.84AAEE280 pKa = 4.0QGEE283 pKa = 4.49LVARR287 pKa = 11.84NAAPTMITRR296 pKa = 11.84TIPAGSCASLEE307 pKa = 4.0QLQDD311 pKa = 3.5ANSDD315 pKa = 3.34KK316 pKa = 11.41AGFQIPVYY324 pKa = 9.49TYY326 pKa = 11.03NPNAGDD332 pKa = 3.87WDD334 pKa = 4.13LLGYY338 pKa = 11.06GDD340 pKa = 4.7VFNAADD346 pKa = 4.23DD347 pKa = 4.74SIVPDD352 pKa = 3.51NQQVFDD358 pKa = 4.68CPADD362 pKa = 3.42NYY364 pKa = 9.86YY365 pKa = 11.12LEE367 pKa = 4.77VVVSNDD373 pKa = 3.81DD374 pKa = 4.56FLRR377 pKa = 11.84DD378 pKa = 3.47WWNLDD383 pKa = 3.31YY384 pKa = 10.87PLLFDD389 pKa = 4.89TPVEE393 pKa = 3.84LCANVKK399 pKa = 9.91LLNPSGNPVAGSYY412 pKa = 10.77VFLSDD417 pKa = 3.43PSGNRR422 pKa = 11.84SFSDD426 pKa = 3.85TYY428 pKa = 11.61AVTDD432 pKa = 4.12GQGSVALSTTLLNPDD447 pKa = 3.03NTQRR451 pKa = 11.84TASLRR456 pKa = 11.84HH457 pKa = 5.86WGYY460 pKa = 11.1QSGTYY465 pKa = 9.34LASDD469 pKa = 3.47IQLSEE474 pKa = 4.09QEE476 pKa = 4.28SCGEE480 pKa = 4.02VQVITVQKK488 pKa = 10.57PDD490 pKa = 3.29MCSVSGTVTRR500 pKa = 11.84AAGGSTDD507 pKa = 4.2EE508 pKa = 4.18NSGWLYY514 pKa = 11.01AFPTNTEE521 pKa = 3.72GYY523 pKa = 8.67MNFGFTQAQANGQYY537 pKa = 11.32SMDD540 pKa = 3.82VEE542 pKa = 4.5CDD544 pKa = 2.65IDD546 pKa = 3.79YY547 pKa = 11.18QFYY550 pKa = 9.56TFSDD554 pKa = 5.17FYY556 pKa = 11.33QGINAEE562 pKa = 4.13QEE564 pKa = 4.16AQAKK568 pKa = 6.6TFNVNGTVSGFEE580 pKa = 4.07TSDD583 pKa = 4.02DD584 pKa = 3.51GDD586 pKa = 3.68KK587 pKa = 11.28AILQDD592 pKa = 5.26AEE594 pKa = 4.24IQNLAPLAILTGHH607 pKa = 7.38IMPQGQSMSFLLMDD621 pKa = 4.55FDD623 pKa = 4.71FDD625 pKa = 4.44YY626 pKa = 10.81PVSYY630 pKa = 10.58RR631 pKa = 11.84LEE633 pKa = 4.04FTDD636 pKa = 3.27MNGAVVASVEE646 pKa = 4.04GTAANEE652 pKa = 3.5IDD654 pKa = 4.34YY655 pKa = 10.94DD656 pKa = 4.27SEE658 pKa = 4.68DD659 pKa = 3.32MGAAPFTLPTDD670 pKa = 4.25LPLGSYY676 pKa = 9.69SVSGILTDD684 pKa = 3.35ATGNQGSASGIIGVTEE700 pKa = 3.81
Molecular weight: 74.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A418YE75|A0A418YE75_9GAMM DUF2867 domain-containing protein OS=Motilimonas pumilua OX=2303987 GN=D1Z90_10935 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.2GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 6.8ATKK25 pKa = 9.91GGRR28 pKa = 11.84QVVARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.48GRR39 pKa = 11.84TRR41 pKa = 11.84LSAA44 pKa = 3.71
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.2GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 6.8ATKK25 pKa = 9.91GGRR28 pKa = 11.84QVVARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.48GRR39 pKa = 11.84TRR41 pKa = 11.84LSAA44 pKa = 3.71
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1321219 |
26 |
4334 |
323.3 |
35.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.251 ± 0.039 | 1.107 ± 0.013 |
5.42 ± 0.039 | 5.821 ± 0.034 |
4.109 ± 0.027 | 6.604 ± 0.028 |
2.377 ± 0.022 | 5.967 ± 0.031 |
5.152 ± 0.035 | 10.684 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.461 ± 0.016 | 4.062 ± 0.026 |
4.076 ± 0.025 | 5.803 ± 0.044 |
4.219 ± 0.025 | 6.528 ± 0.031 |
5.147 ± 0.032 | 6.834 ± 0.031 |
1.32 ± 0.018 | 3.059 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
