
candidate division SR1 bacterium RAAC1_SR1_1
Taxonomy: cellular organisms; Bacteria; Bacteria incertae sedis; Bacteria candidate phyla; Candidatus Absconditabacteria; unclassified Candidatus Absconditabacteria
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
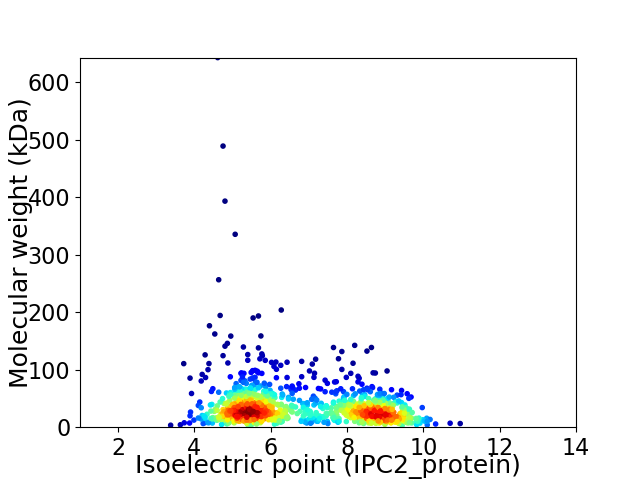
Virtual 2D-PAGE plot for 1059 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A288DD64|A0A288DD64_9BACT Uncharacterized protein OS=candidate division SR1 bacterium RAAC1_SR1_1 OX=1394709 GN=P148_SR1C00001G0750 PE=4 SV=1
MM1 pKa = 7.4KK2 pKa = 10.22KK3 pKa = 9.8ILIAALFILGIFASFTRR20 pKa = 11.84AEE22 pKa = 4.28EE23 pKa = 4.18LNSGNVISTFNCEE36 pKa = 3.81TGATSCDD43 pKa = 3.14LFGRR47 pKa = 11.84NITSIASDD55 pKa = 3.34SFINHH60 pKa = 6.28SNLQYY65 pKa = 11.01LAIRR69 pKa = 11.84NNPLTGIDD77 pKa = 3.24VGDD80 pKa = 4.01FVGLDD85 pKa = 3.65NLIGLYY91 pKa = 9.39MDD93 pKa = 4.87GGNLTSIEE101 pKa = 4.25SGSFLGLSNLNIIDD115 pKa = 4.56LDD117 pKa = 4.01QNKK120 pKa = 8.16LTTIKK125 pKa = 10.75AGTFNGLSSLIRR137 pKa = 11.84LEE139 pKa = 3.95ISQNQITSIEE149 pKa = 4.19SGGFLGATNLTGLLLDD165 pKa = 4.3NNLITGIDD173 pKa = 3.35VGDD176 pKa = 4.46FIGLTNLDD184 pKa = 4.29YY185 pKa = 11.47LSLTQNQISSIEE197 pKa = 4.03RR198 pKa = 11.84GSFSGLFLLKK208 pKa = 10.41NLYY211 pKa = 10.63LNYY214 pKa = 10.56NQITSIDD221 pKa = 3.54VGDD224 pKa = 5.56FIGLDD229 pKa = 3.24NLTQLNLNINKK240 pKa = 9.41ISSIEE245 pKa = 3.89AYY247 pKa = 10.05GFSEE251 pKa = 4.7LSSLTYY257 pKa = 11.04LNLGYY262 pKa = 10.39NQINSIEE269 pKa = 4.21SGSFYY274 pKa = 11.15GLDD277 pKa = 3.55NLTDD281 pKa = 4.37LYY283 pKa = 11.18LGNFKK288 pKa = 10.56LPRR291 pKa = 11.84DD292 pKa = 3.69IGPEE296 pKa = 3.72NVVYY300 pKa = 10.61NQISSIDD307 pKa = 3.54VGDD310 pKa = 3.75FAGLNNLTQLSLDD323 pKa = 3.6GNAISSIEE331 pKa = 4.06SGSFIGLSFLTYY343 pKa = 10.75LDD345 pKa = 4.58LNNNQIDD352 pKa = 4.34SIDD355 pKa = 3.95LGDD358 pKa = 4.11FADD361 pKa = 5.03LSSLTDD367 pKa = 3.95LYY369 pKa = 11.34LYY371 pKa = 10.86GNEE374 pKa = 3.66ISFIEE379 pKa = 4.37SNSFNLLSSLTYY391 pKa = 10.66LDD393 pKa = 4.97LGNNQINSIDD403 pKa = 3.23IGDD406 pKa = 3.95FVGLTNLTDD415 pKa = 4.14LYY417 pKa = 11.03LYY419 pKa = 10.75GNEE422 pKa = 3.87ISSIGSNSFNLLSSLTYY439 pKa = 10.73LDD441 pKa = 5.17LNSNQINSIEE451 pKa = 4.15SGDD454 pKa = 3.9FNGLSSLIFLDD465 pKa = 5.3LSTNQINSIEE475 pKa = 4.19SGDD478 pKa = 3.9FNGLSSLNEE487 pKa = 4.07LYY489 pKa = 10.88LSYY492 pKa = 11.11NQINSIEE499 pKa = 4.01VSGFSGLPSLMYY511 pKa = 10.57LYY513 pKa = 10.87LDD515 pKa = 3.75NNQLLSIDD523 pKa = 3.38VGNFYY528 pKa = 10.86EE529 pKa = 4.9LDD531 pKa = 3.72NLNGLSLNKK540 pKa = 10.19NFISSITSNVFDD552 pKa = 4.5NLLNLWDD559 pKa = 4.83LSLSSICTDD568 pKa = 3.0NPGIIAYY575 pKa = 8.88PNGSTEE581 pKa = 4.18TNNNYY586 pKa = 10.05CSVINSTNQIDD597 pKa = 5.41FITEE601 pKa = 3.79LTKK604 pKa = 10.84EE605 pKa = 4.18VMDD608 pKa = 4.72GKK610 pKa = 9.33ITLALYY616 pKa = 10.32SLSGNYY622 pKa = 7.46YY623 pKa = 7.88TGTYY627 pKa = 9.82AISFTPGITNEE638 pKa = 3.97ILVDD642 pKa = 4.61IIMQSPTSNYY652 pKa = 8.05TQDD655 pKa = 3.52YY656 pKa = 8.3YY657 pKa = 11.78LNNVEE662 pKa = 4.11VQYY665 pKa = 11.58DD666 pKa = 3.79SGTLLTTNGQGFTGILQAPVLLSTGSTKK694 pKa = 10.75LVDD697 pKa = 3.46NVVALTKK704 pKa = 10.84FGDD707 pKa = 3.25ATKK710 pKa = 10.49RR711 pKa = 11.84IDD713 pKa = 3.51FSKK716 pKa = 10.63SVTVRR721 pKa = 11.84MPALGKK727 pKa = 10.5NIDD730 pKa = 3.9DD731 pKa = 4.08VVDD734 pKa = 3.74IFYY737 pKa = 10.98SQNLSSNWSFHH748 pKa = 4.98GTSTVIDD755 pKa = 3.59IAGIPYY761 pKa = 9.83VEE763 pKa = 4.69FTTDD767 pKa = 2.61HH768 pKa = 5.96ATYY771 pKa = 9.83FAIGEE776 pKa = 4.47GTGSFVINNDD786 pKa = 3.62DD787 pKa = 4.75ASTNSQNVTLNISGAIAYY805 pKa = 7.51MRR807 pKa = 11.84FSNDD811 pKa = 2.57NSNRR815 pKa = 11.84SAWEE819 pKa = 4.17SISDD823 pKa = 3.71TKK825 pKa = 11.29SRR827 pKa = 11.84TLSAGTGTKK836 pKa = 8.43TVYY839 pKa = 10.95AEE841 pKa = 4.31FDD843 pKa = 3.66LDD845 pKa = 3.67SDD847 pKa = 4.62FIADD851 pKa = 3.73ISTSDD856 pKa = 3.52TIEE859 pKa = 4.05YY860 pKa = 10.13TDD862 pKa = 3.87GSSGNGTNGNITLTITGGVTEE883 pKa = 5.18CIYY886 pKa = 10.08GTSLTMDD893 pKa = 3.43AQEE896 pKa = 4.24VKK898 pKa = 10.45IGVPYY903 pKa = 10.3TFTGTFPSTWYY914 pKa = 9.58CQDD917 pKa = 2.89YY918 pKa = 10.97RR919 pKa = 11.84GINGGWTLTIQTTDD933 pKa = 4.5LINEE937 pKa = 4.13KK938 pKa = 10.79SNVISGSNLLISHH951 pKa = 6.44NPVVVEE957 pKa = 4.31GDD959 pKa = 3.86SACTGDD965 pKa = 5.24NGTATQFYY973 pKa = 8.85SAPYY977 pKa = 9.63EE978 pKa = 4.08IFEE981 pKa = 5.7KK982 pKa = 10.63IPDD985 pKa = 3.65SNKK988 pKa = 8.87ICRR991 pKa = 11.84VSADD995 pKa = 3.49NVGLLVNVPANQAPGSYY1012 pKa = 10.0SGTLTLTMNGFF1023 pKa = 3.76
MM1 pKa = 7.4KK2 pKa = 10.22KK3 pKa = 9.8ILIAALFILGIFASFTRR20 pKa = 11.84AEE22 pKa = 4.28EE23 pKa = 4.18LNSGNVISTFNCEE36 pKa = 3.81TGATSCDD43 pKa = 3.14LFGRR47 pKa = 11.84NITSIASDD55 pKa = 3.34SFINHH60 pKa = 6.28SNLQYY65 pKa = 11.01LAIRR69 pKa = 11.84NNPLTGIDD77 pKa = 3.24VGDD80 pKa = 4.01FVGLDD85 pKa = 3.65NLIGLYY91 pKa = 9.39MDD93 pKa = 4.87GGNLTSIEE101 pKa = 4.25SGSFLGLSNLNIIDD115 pKa = 4.56LDD117 pKa = 4.01QNKK120 pKa = 8.16LTTIKK125 pKa = 10.75AGTFNGLSSLIRR137 pKa = 11.84LEE139 pKa = 3.95ISQNQITSIEE149 pKa = 4.19SGGFLGATNLTGLLLDD165 pKa = 4.3NNLITGIDD173 pKa = 3.35VGDD176 pKa = 4.46FIGLTNLDD184 pKa = 4.29YY185 pKa = 11.47LSLTQNQISSIEE197 pKa = 4.03RR198 pKa = 11.84GSFSGLFLLKK208 pKa = 10.41NLYY211 pKa = 10.63LNYY214 pKa = 10.56NQITSIDD221 pKa = 3.54VGDD224 pKa = 5.56FIGLDD229 pKa = 3.24NLTQLNLNINKK240 pKa = 9.41ISSIEE245 pKa = 3.89AYY247 pKa = 10.05GFSEE251 pKa = 4.7LSSLTYY257 pKa = 11.04LNLGYY262 pKa = 10.39NQINSIEE269 pKa = 4.21SGSFYY274 pKa = 11.15GLDD277 pKa = 3.55NLTDD281 pKa = 4.37LYY283 pKa = 11.18LGNFKK288 pKa = 10.56LPRR291 pKa = 11.84DD292 pKa = 3.69IGPEE296 pKa = 3.72NVVYY300 pKa = 10.61NQISSIDD307 pKa = 3.54VGDD310 pKa = 3.75FAGLNNLTQLSLDD323 pKa = 3.6GNAISSIEE331 pKa = 4.06SGSFIGLSFLTYY343 pKa = 10.75LDD345 pKa = 4.58LNNNQIDD352 pKa = 4.34SIDD355 pKa = 3.95LGDD358 pKa = 4.11FADD361 pKa = 5.03LSSLTDD367 pKa = 3.95LYY369 pKa = 11.34LYY371 pKa = 10.86GNEE374 pKa = 3.66ISFIEE379 pKa = 4.37SNSFNLLSSLTYY391 pKa = 10.66LDD393 pKa = 4.97LGNNQINSIDD403 pKa = 3.23IGDD406 pKa = 3.95FVGLTNLTDD415 pKa = 4.14LYY417 pKa = 11.03LYY419 pKa = 10.75GNEE422 pKa = 3.87ISSIGSNSFNLLSSLTYY439 pKa = 10.73LDD441 pKa = 5.17LNSNQINSIEE451 pKa = 4.15SGDD454 pKa = 3.9FNGLSSLIFLDD465 pKa = 5.3LSTNQINSIEE475 pKa = 4.19SGDD478 pKa = 3.9FNGLSSLNEE487 pKa = 4.07LYY489 pKa = 10.88LSYY492 pKa = 11.11NQINSIEE499 pKa = 4.01VSGFSGLPSLMYY511 pKa = 10.57LYY513 pKa = 10.87LDD515 pKa = 3.75NNQLLSIDD523 pKa = 3.38VGNFYY528 pKa = 10.86EE529 pKa = 4.9LDD531 pKa = 3.72NLNGLSLNKK540 pKa = 10.19NFISSITSNVFDD552 pKa = 4.5NLLNLWDD559 pKa = 4.83LSLSSICTDD568 pKa = 3.0NPGIIAYY575 pKa = 8.88PNGSTEE581 pKa = 4.18TNNNYY586 pKa = 10.05CSVINSTNQIDD597 pKa = 5.41FITEE601 pKa = 3.79LTKK604 pKa = 10.84EE605 pKa = 4.18VMDD608 pKa = 4.72GKK610 pKa = 9.33ITLALYY616 pKa = 10.32SLSGNYY622 pKa = 7.46YY623 pKa = 7.88TGTYY627 pKa = 9.82AISFTPGITNEE638 pKa = 3.97ILVDD642 pKa = 4.61IIMQSPTSNYY652 pKa = 8.05TQDD655 pKa = 3.52YY656 pKa = 8.3YY657 pKa = 11.78LNNVEE662 pKa = 4.11VQYY665 pKa = 11.58DD666 pKa = 3.79SGTLLTTNGQGFTGILQAPVLLSTGSTKK694 pKa = 10.75LVDD697 pKa = 3.46NVVALTKK704 pKa = 10.84FGDD707 pKa = 3.25ATKK710 pKa = 10.49RR711 pKa = 11.84IDD713 pKa = 3.51FSKK716 pKa = 10.63SVTVRR721 pKa = 11.84MPALGKK727 pKa = 10.5NIDD730 pKa = 3.9DD731 pKa = 4.08VVDD734 pKa = 3.74IFYY737 pKa = 10.98SQNLSSNWSFHH748 pKa = 4.98GTSTVIDD755 pKa = 3.59IAGIPYY761 pKa = 9.83VEE763 pKa = 4.69FTTDD767 pKa = 2.61HH768 pKa = 5.96ATYY771 pKa = 9.83FAIGEE776 pKa = 4.47GTGSFVINNDD786 pKa = 3.62DD787 pKa = 4.75ASTNSQNVTLNISGAIAYY805 pKa = 7.51MRR807 pKa = 11.84FSNDD811 pKa = 2.57NSNRR815 pKa = 11.84SAWEE819 pKa = 4.17SISDD823 pKa = 3.71TKK825 pKa = 11.29SRR827 pKa = 11.84TLSAGTGTKK836 pKa = 8.43TVYY839 pKa = 10.95AEE841 pKa = 4.31FDD843 pKa = 3.66LDD845 pKa = 3.67SDD847 pKa = 4.62FIADD851 pKa = 3.73ISTSDD856 pKa = 3.52TIEE859 pKa = 4.05YY860 pKa = 10.13TDD862 pKa = 3.87GSSGNGTNGNITLTITGGVTEE883 pKa = 5.18CIYY886 pKa = 10.08GTSLTMDD893 pKa = 3.43AQEE896 pKa = 4.24VKK898 pKa = 10.45IGVPYY903 pKa = 10.3TFTGTFPSTWYY914 pKa = 9.58CQDD917 pKa = 2.89YY918 pKa = 10.97RR919 pKa = 11.84GINGGWTLTIQTTDD933 pKa = 4.5LINEE937 pKa = 4.13KK938 pKa = 10.79SNVISGSNLLISHH951 pKa = 6.44NPVVVEE957 pKa = 4.31GDD959 pKa = 3.86SACTGDD965 pKa = 5.24NGTATQFYY973 pKa = 8.85SAPYY977 pKa = 9.63EE978 pKa = 4.08IFEE981 pKa = 5.7KK982 pKa = 10.63IPDD985 pKa = 3.65SNKK988 pKa = 8.87ICRR991 pKa = 11.84VSADD995 pKa = 3.49NVGLLVNVPANQAPGSYY1012 pKa = 10.0SGTLTLTMNGFF1023 pKa = 3.76
Molecular weight: 110.61 kDa
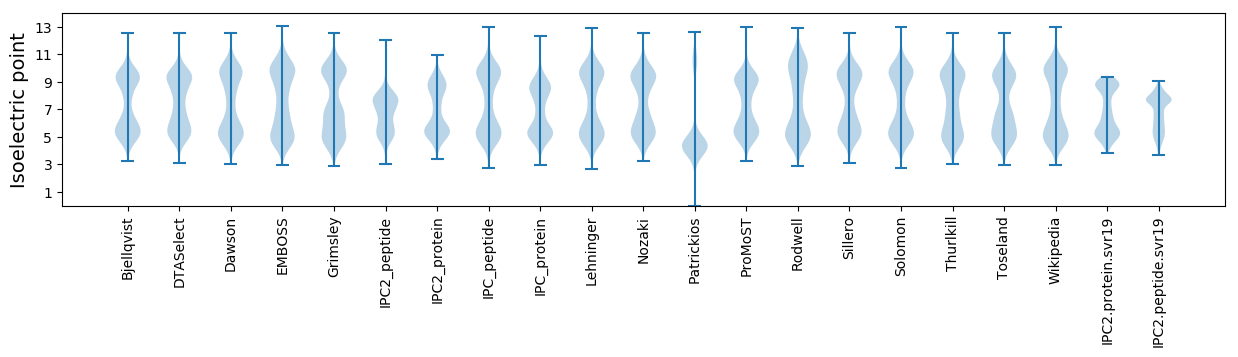
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A288DJ48|A0A288DJ48_9BACT SsrA-binding protein OS=candidate division SR1 bacterium RAAC1_SR1_1 OX=1394709 GN=smpB PE=3 SV=1
MM1 pKa = 7.46ASKK4 pKa = 10.42LRR6 pKa = 11.84RR7 pKa = 11.84VRR9 pKa = 11.84KK10 pKa = 7.87YY11 pKa = 10.4SRR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 9.7RR16 pKa = 11.84LTKK19 pKa = 10.01HH20 pKa = 5.54GFRR23 pKa = 11.84EE24 pKa = 3.94RR25 pKa = 11.84LQTKK29 pKa = 9.65NGKK32 pKa = 9.16KK33 pKa = 9.82LLARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84AKK41 pKa = 10.06GRR43 pKa = 11.84KK44 pKa = 8.83SLTVQRR50 pKa = 11.84KK51 pKa = 7.82KK52 pKa = 11.02
MM1 pKa = 7.46ASKK4 pKa = 10.42LRR6 pKa = 11.84RR7 pKa = 11.84VRR9 pKa = 11.84KK10 pKa = 7.87YY11 pKa = 10.4SRR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 9.7RR16 pKa = 11.84LTKK19 pKa = 10.01HH20 pKa = 5.54GFRR23 pKa = 11.84EE24 pKa = 3.94RR25 pKa = 11.84LQTKK29 pKa = 9.65NGKK32 pKa = 9.16KK33 pKa = 9.82LLARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84AKK41 pKa = 10.06GRR43 pKa = 11.84KK44 pKa = 8.83SLTVQRR50 pKa = 11.84KK51 pKa = 7.82KK52 pKa = 11.02
Molecular weight: 6.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
355954 |
29 |
5926 |
336.1 |
37.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.092 ± 0.066 | 0.936 ± 0.038 |
5.48 ± 0.064 | 6.591 ± 0.117 |
5.014 ± 0.068 | 6.765 ± 0.102 |
1.595 ± 0.048 | 9.289 ± 0.088 |
8.726 ± 0.164 | 9.073 ± 0.095 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.185 ± 0.049 | 5.869 ± 0.147 |
3.156 ± 0.049 | 3.99 ± 0.052 |
3.194 ± 0.059 | 6.393 ± 0.122 |
6.294 ± 0.173 | 5.535 ± 0.075 |
0.705 ± 0.019 | 4.119 ± 0.063 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
