
Paenibacillus methanolicus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Paenibacillaceae; Paenibacillus
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
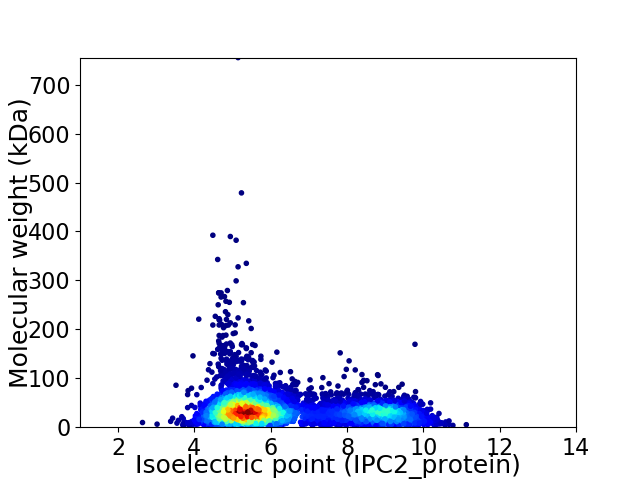
Virtual 2D-PAGE plot for 6989 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5S5C6Y4|A0A5S5C6Y4_9BACL Diguanylate cyclase (GGDEF)-like protein OS=Paenibacillus methanolicus OX=582686 GN=BCM02_106438 PE=4 SV=1
MM1 pKa = 7.45QGLFVLNTRR10 pKa = 11.84TIGRR14 pKa = 11.84KK15 pKa = 9.08IGAALLAASLALPGSVIYY33 pKa = 10.77AEE35 pKa = 5.39EE36 pKa = 5.37GDD38 pKa = 4.5DD39 pKa = 4.84WIPDD43 pKa = 3.55PPAAISFDD51 pKa = 3.83APVVTVDD58 pKa = 3.34EE59 pKa = 4.75GAGYY63 pKa = 10.62AYY65 pKa = 10.63VGVSRR70 pKa = 11.84TGAASSLTVDD80 pKa = 3.31YY81 pKa = 8.8ATYY84 pKa = 10.76GGTATEE90 pKa = 4.31GTDD93 pKa = 3.32YY94 pKa = 11.11LAASGQITFGPGEE107 pKa = 4.08TFATIEE113 pKa = 4.01IPITNDD119 pKa = 2.56IQFEE123 pKa = 4.12PAEE126 pKa = 4.32SFSVEE131 pKa = 4.22LFNVSDD137 pKa = 4.13GVTSIDD143 pKa = 3.52RR144 pKa = 11.84GTTTVTIADD153 pKa = 4.09NDD155 pKa = 3.84PDD157 pKa = 3.81VPYY160 pKa = 11.21NPGKK164 pKa = 10.7LGFATALTEE173 pKa = 3.95VDD175 pKa = 3.98EE176 pKa = 4.2NHH178 pKa = 7.39PDD180 pKa = 3.37GTVSLTVARR189 pKa = 11.84SEE191 pKa = 4.2GTDD194 pKa = 3.44GYY196 pKa = 11.44VAVDD200 pKa = 3.41YY201 pKa = 11.27QIDD204 pKa = 3.76GGTAGNGADD213 pKa = 4.0FEE215 pKa = 4.97GTGGTLSFEE224 pKa = 4.7PGEE227 pKa = 4.19SSKK230 pKa = 9.96TIQVTLYY237 pKa = 10.9DD238 pKa = 4.04DD239 pKa = 4.77AEE241 pKa = 4.66SEE243 pKa = 4.36GNEE246 pKa = 4.04YY247 pKa = 10.74FHH249 pKa = 7.33LSLSNPTNDD258 pKa = 2.83ATLAEE263 pKa = 4.34SSFTKK268 pKa = 10.45VRR270 pKa = 11.84ILDD273 pKa = 3.77NEE275 pKa = 3.98PWTPRR280 pKa = 11.84AGSFEE285 pKa = 4.53VNPTAPMGEE294 pKa = 3.99ASGTHH299 pKa = 6.19YY300 pKa = 10.96INVIRR305 pKa = 11.84TDD307 pKa = 3.56GADD310 pKa = 3.04VEE312 pKa = 4.54AAIDD316 pKa = 3.67YY317 pKa = 7.62TIAGVTAEE325 pKa = 4.63EE326 pKa = 4.99GSDD329 pKa = 3.58FTGSSGTLVFPVDD342 pKa = 3.48EE343 pKa = 4.23TLQYY347 pKa = 10.66IPLPILDD354 pKa = 4.7DD355 pKa = 4.25ALSEE359 pKa = 4.29GNEE362 pKa = 4.21TLTITLSNPTSGAVIGTYY380 pKa = 7.19GTRR383 pKa = 11.84TYY385 pKa = 10.19TIEE388 pKa = 4.42DD389 pKa = 3.85NEE391 pKa = 4.19PHH393 pKa = 6.63EE394 pKa = 4.39EE395 pKa = 3.89QHH397 pKa = 6.77PGMFMLGASQYY408 pKa = 11.5AVDD411 pKa = 4.36EE412 pKa = 4.66NGGSAKK418 pKa = 10.58LQVTRR423 pKa = 11.84MDD425 pKa = 3.92GADD428 pKa = 3.54GEE430 pKa = 4.59ATLTYY435 pKa = 10.42SILGGSAAEE444 pKa = 4.12NADD447 pKa = 3.44YY448 pKa = 11.08AGNSGQLTFADD459 pKa = 4.32GEE461 pKa = 4.51TEE463 pKa = 4.16KK464 pKa = 10.9FIEE467 pKa = 4.29VPIVDD472 pKa = 4.58DD473 pKa = 3.93SMHH476 pKa = 5.89EE477 pKa = 4.05AEE479 pKa = 4.7EE480 pKa = 4.07QFQVRR485 pKa = 11.84LLDD488 pKa = 3.61ATGGAVVGPNYY499 pKa = 10.48LSGVTIKK506 pKa = 11.1DD507 pKa = 3.28NDD509 pKa = 4.13PAGVLQFSPFTTTVYY524 pKa = 10.66EE525 pKa = 4.06EE526 pKa = 4.4SKK528 pKa = 10.5QVTMTVARR536 pKa = 11.84TGDD539 pKa = 3.42TSGTASVAYY548 pKa = 8.24ATKK551 pKa = 10.22DD552 pKa = 3.44YY553 pKa = 11.11SALAGRR559 pKa = 11.84DD560 pKa = 3.34YY561 pKa = 11.2AAQSGVLTFAPGEE574 pKa = 4.25TQKK577 pKa = 11.23KK578 pKa = 7.35ITITMYY584 pKa = 10.93DD585 pKa = 3.66DD586 pKa = 4.47SVKK589 pKa = 10.48EE590 pKa = 3.75WFEE593 pKa = 3.92YY594 pKa = 10.6FSILLSNPAGAEE606 pKa = 3.77LGTYY610 pKa = 8.19STGLVYY616 pKa = 10.81VYY618 pKa = 11.11DD619 pKa = 4.01NDD621 pKa = 3.86KK622 pKa = 9.63YY623 pKa = 9.46TYY625 pKa = 7.31PTWPRR630 pKa = 11.84YY631 pKa = 9.0
MM1 pKa = 7.45QGLFVLNTRR10 pKa = 11.84TIGRR14 pKa = 11.84KK15 pKa = 9.08IGAALLAASLALPGSVIYY33 pKa = 10.77AEE35 pKa = 5.39EE36 pKa = 5.37GDD38 pKa = 4.5DD39 pKa = 4.84WIPDD43 pKa = 3.55PPAAISFDD51 pKa = 3.83APVVTVDD58 pKa = 3.34EE59 pKa = 4.75GAGYY63 pKa = 10.62AYY65 pKa = 10.63VGVSRR70 pKa = 11.84TGAASSLTVDD80 pKa = 3.31YY81 pKa = 8.8ATYY84 pKa = 10.76GGTATEE90 pKa = 4.31GTDD93 pKa = 3.32YY94 pKa = 11.11LAASGQITFGPGEE107 pKa = 4.08TFATIEE113 pKa = 4.01IPITNDD119 pKa = 2.56IQFEE123 pKa = 4.12PAEE126 pKa = 4.32SFSVEE131 pKa = 4.22LFNVSDD137 pKa = 4.13GVTSIDD143 pKa = 3.52RR144 pKa = 11.84GTTTVTIADD153 pKa = 4.09NDD155 pKa = 3.84PDD157 pKa = 3.81VPYY160 pKa = 11.21NPGKK164 pKa = 10.7LGFATALTEE173 pKa = 3.95VDD175 pKa = 3.98EE176 pKa = 4.2NHH178 pKa = 7.39PDD180 pKa = 3.37GTVSLTVARR189 pKa = 11.84SEE191 pKa = 4.2GTDD194 pKa = 3.44GYY196 pKa = 11.44VAVDD200 pKa = 3.41YY201 pKa = 11.27QIDD204 pKa = 3.76GGTAGNGADD213 pKa = 4.0FEE215 pKa = 4.97GTGGTLSFEE224 pKa = 4.7PGEE227 pKa = 4.19SSKK230 pKa = 9.96TIQVTLYY237 pKa = 10.9DD238 pKa = 4.04DD239 pKa = 4.77AEE241 pKa = 4.66SEE243 pKa = 4.36GNEE246 pKa = 4.04YY247 pKa = 10.74FHH249 pKa = 7.33LSLSNPTNDD258 pKa = 2.83ATLAEE263 pKa = 4.34SSFTKK268 pKa = 10.45VRR270 pKa = 11.84ILDD273 pKa = 3.77NEE275 pKa = 3.98PWTPRR280 pKa = 11.84AGSFEE285 pKa = 4.53VNPTAPMGEE294 pKa = 3.99ASGTHH299 pKa = 6.19YY300 pKa = 10.96INVIRR305 pKa = 11.84TDD307 pKa = 3.56GADD310 pKa = 3.04VEE312 pKa = 4.54AAIDD316 pKa = 3.67YY317 pKa = 7.62TIAGVTAEE325 pKa = 4.63EE326 pKa = 4.99GSDD329 pKa = 3.58FTGSSGTLVFPVDD342 pKa = 3.48EE343 pKa = 4.23TLQYY347 pKa = 10.66IPLPILDD354 pKa = 4.7DD355 pKa = 4.25ALSEE359 pKa = 4.29GNEE362 pKa = 4.21TLTITLSNPTSGAVIGTYY380 pKa = 7.19GTRR383 pKa = 11.84TYY385 pKa = 10.19TIEE388 pKa = 4.42DD389 pKa = 3.85NEE391 pKa = 4.19PHH393 pKa = 6.63EE394 pKa = 4.39EE395 pKa = 3.89QHH397 pKa = 6.77PGMFMLGASQYY408 pKa = 11.5AVDD411 pKa = 4.36EE412 pKa = 4.66NGGSAKK418 pKa = 10.58LQVTRR423 pKa = 11.84MDD425 pKa = 3.92GADD428 pKa = 3.54GEE430 pKa = 4.59ATLTYY435 pKa = 10.42SILGGSAAEE444 pKa = 4.12NADD447 pKa = 3.44YY448 pKa = 11.08AGNSGQLTFADD459 pKa = 4.32GEE461 pKa = 4.51TEE463 pKa = 4.16KK464 pKa = 10.9FIEE467 pKa = 4.29VPIVDD472 pKa = 4.58DD473 pKa = 3.93SMHH476 pKa = 5.89EE477 pKa = 4.05AEE479 pKa = 4.7EE480 pKa = 4.07QFQVRR485 pKa = 11.84LLDD488 pKa = 3.61ATGGAVVGPNYY499 pKa = 10.48LSGVTIKK506 pKa = 11.1DD507 pKa = 3.28NDD509 pKa = 4.13PAGVLQFSPFTTTVYY524 pKa = 10.66EE525 pKa = 4.06EE526 pKa = 4.4SKK528 pKa = 10.5QVTMTVARR536 pKa = 11.84TGDD539 pKa = 3.42TSGTASVAYY548 pKa = 8.24ATKK551 pKa = 10.22DD552 pKa = 3.44YY553 pKa = 11.11SALAGRR559 pKa = 11.84DD560 pKa = 3.34YY561 pKa = 11.2AAQSGVLTFAPGEE574 pKa = 4.25TQKK577 pKa = 11.23KK578 pKa = 7.35ITITMYY584 pKa = 10.93DD585 pKa = 3.66DD586 pKa = 4.47SVKK589 pKa = 10.48EE590 pKa = 3.75WFEE593 pKa = 3.92YY594 pKa = 10.6FSILLSNPAGAEE606 pKa = 3.77LGTYY610 pKa = 8.19STGLVYY616 pKa = 10.81VYY618 pKa = 11.11DD619 pKa = 4.01NDD621 pKa = 3.86KK622 pKa = 9.63YY623 pKa = 9.46TYY625 pKa = 7.31PTWPRR630 pKa = 11.84YY631 pKa = 9.0
Molecular weight: 66.67 kDa
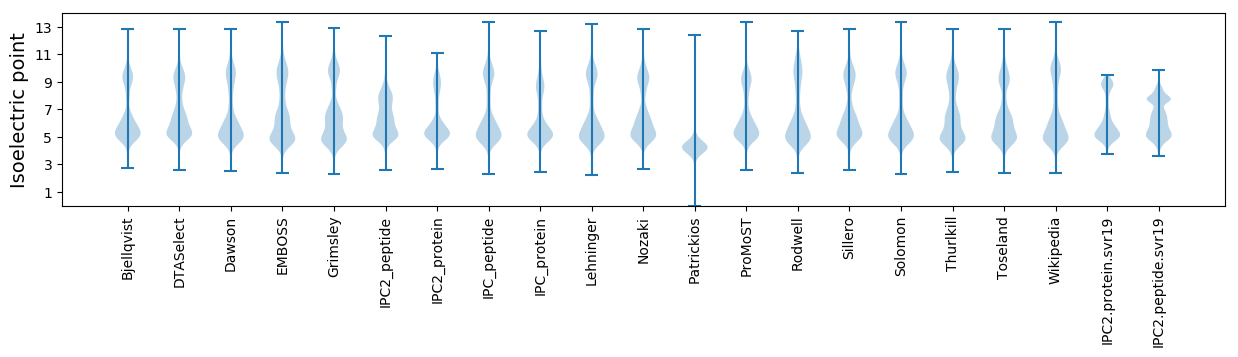
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5S5C7S6|A0A5S5C7S6_9BACL Putative NADPH-quinone reductase OS=Paenibacillus methanolicus OX=582686 GN=BCM02_106324 PE=4 SV=1
MM1 pKa = 7.9RR2 pKa = 11.84PTFKK6 pKa = 10.82PNVSKK11 pKa = 10.8RR12 pKa = 11.84KK13 pKa = 8.95KK14 pKa = 8.25VHH16 pKa = 5.48GFRR19 pKa = 11.84KK20 pKa = 10.06RR21 pKa = 11.84MASKK25 pKa = 10.64NGRR28 pKa = 11.84KK29 pKa = 9.12VLAARR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.23GRR39 pKa = 11.84KK40 pKa = 8.56VLSAA44 pKa = 4.05
MM1 pKa = 7.9RR2 pKa = 11.84PTFKK6 pKa = 10.82PNVSKK11 pKa = 10.8RR12 pKa = 11.84KK13 pKa = 8.95KK14 pKa = 8.25VHH16 pKa = 5.48GFRR19 pKa = 11.84KK20 pKa = 10.06RR21 pKa = 11.84MASKK25 pKa = 10.64NGRR28 pKa = 11.84KK29 pKa = 9.12VLAARR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.23GRR39 pKa = 11.84KK40 pKa = 8.56VLSAA44 pKa = 4.05
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2386252 |
29 |
6708 |
341.4 |
37.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.137 ± 0.035 | 0.687 ± 0.008 |
5.281 ± 0.023 | 6.551 ± 0.035 |
3.984 ± 0.023 | 7.977 ± 0.03 |
2.002 ± 0.013 | 5.905 ± 0.026 |
4.457 ± 0.028 | 9.97 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.747 ± 0.015 | 3.474 ± 0.023 |
4.239 ± 0.019 | 3.603 ± 0.017 |
5.79 ± 0.033 | 5.943 ± 0.023 |
5.43 ± 0.027 | 6.998 ± 0.023 |
1.38 ± 0.013 | 3.445 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
