
Arthrobacter subterraneus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Micrococcaceae; Arthrobacter
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
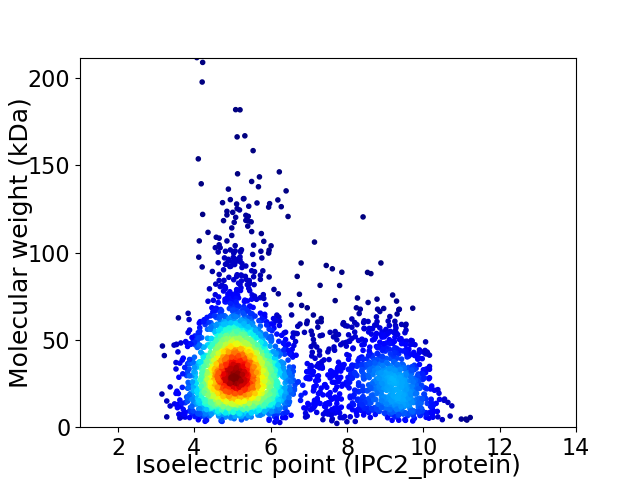
Virtual 2D-PAGE plot for 3624 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G8KK19|A0A1G8KK19_9MICC Succinate semialdehyde dehydrogenase OS=Arthrobacter subterraneus OX=335973 GN=SAMN04488693_11168 PE=3 SV=1
MM1 pKa = 6.79THH3 pKa = 5.84SHH5 pKa = 6.74PARR8 pKa = 11.84RR9 pKa = 11.84SPLRR13 pKa = 11.84ALAGAAAVVVGLVLSGCTAGPDD35 pKa = 3.54TGAVTAPPEE44 pKa = 4.35VIGEE48 pKa = 4.16VPEE51 pKa = 4.29DD52 pKa = 3.42LRR54 pKa = 11.84EE55 pKa = 4.19FYY57 pKa = 10.75AQEE60 pKa = 4.27VTWEE64 pKa = 4.12EE65 pKa = 4.54CEE67 pKa = 4.47GDD69 pKa = 4.32DD70 pKa = 5.19FACATVKK77 pKa = 10.57VPLDD81 pKa = 3.45YY82 pKa = 11.03SDD84 pKa = 4.6PGRR87 pKa = 11.84ASIDD91 pKa = 2.9IAMIRR96 pKa = 11.84LDD98 pKa = 4.4APGDD102 pKa = 3.81AQGSLLVNPGGPGVSGYY119 pKa = 11.17NLVRR123 pKa = 11.84DD124 pKa = 3.72SAAFILSDD132 pKa = 4.09RR133 pKa = 11.84LRR135 pKa = 11.84QSFDD139 pKa = 3.18VVGFDD144 pKa = 3.37PRR146 pKa = 11.84GVNEE150 pKa = 4.16SSAVEE155 pKa = 4.1CLTDD159 pKa = 3.48EE160 pKa = 4.52EE161 pKa = 4.67RR162 pKa = 11.84DD163 pKa = 3.4EE164 pKa = 5.03ARR166 pKa = 11.84AEE168 pKa = 4.11TLPPDD173 pKa = 3.87ATDD176 pKa = 4.06AEE178 pKa = 4.21ALEE181 pKa = 4.19ILEE184 pKa = 4.79ADD186 pKa = 3.53SAEE189 pKa = 4.14YY190 pKa = 10.51AQLCAEE196 pKa = 4.32RR197 pKa = 11.84TGEE200 pKa = 4.09LLGHH204 pKa = 7.11IDD206 pKa = 3.58TVSAAKK212 pKa = 10.9DD213 pKa = 3.4MDD215 pKa = 3.52ILRR218 pKa = 11.84AVLGDD223 pKa = 3.5EE224 pKa = 3.91QLNYY228 pKa = 10.92LGFSYY233 pKa = 8.74GTQLGAAYY241 pKa = 10.26AGLFPGNVGRR251 pKa = 11.84FVLDD255 pKa = 3.69GAVDD259 pKa = 4.12PSLSSAEE266 pKa = 3.8ITAGQVAAFDD276 pKa = 3.67EE277 pKa = 4.39ALRR280 pKa = 11.84NYY282 pKa = 9.87VADD285 pKa = 3.99CQSQSEE291 pKa = 4.67CPLPGNVDD299 pKa = 3.18EE300 pKa = 5.38GVAVVDD306 pKa = 3.87EE307 pKa = 4.57LFEE310 pKa = 4.39SVEE313 pKa = 4.14EE314 pKa = 4.43SPMTAADD321 pKa = 3.92GRR323 pKa = 11.84PVTIGTFFQGFVLPLYY339 pKa = 10.75DD340 pKa = 3.69SASWPALTAAIDD352 pKa = 3.45QALQGDD358 pKa = 4.0PSEE361 pKa = 4.52FLRR364 pKa = 11.84FADD367 pKa = 4.44LGADD371 pKa = 3.85RR372 pKa = 11.84SPTGDD377 pKa = 3.56YY378 pKa = 8.55LTNSAAAFTAVTCLDD393 pKa = 3.85YY394 pKa = 11.23PMPADD399 pKa = 3.98PGVMAEE405 pKa = 3.97EE406 pKa = 4.59AEE408 pKa = 4.09ALEE411 pKa = 4.44ASSAVFGEE419 pKa = 4.12YY420 pKa = 10.4LSYY423 pKa = 11.15GALACKK429 pKa = 9.69NWAYY433 pKa = 10.52EE434 pKa = 4.13PVNEE438 pKa = 4.7PSPASAPGAPPMLVIGTTGDD458 pKa = 3.36PATPYY463 pKa = 9.88EE464 pKa = 4.19WSLALAEE471 pKa = 4.11QLEE474 pKa = 4.9SAVHH478 pKa = 4.68VTWEE482 pKa = 4.56GQGHH486 pKa = 5.01TAYY489 pKa = 10.14GRR491 pKa = 11.84SGEE494 pKa = 4.41CIEE497 pKa = 4.92SLVDD501 pKa = 4.77DD502 pKa = 4.49FLIDD506 pKa = 3.45GTVPEE511 pKa = 4.47MSARR515 pKa = 11.84CC516 pKa = 3.57
MM1 pKa = 6.79THH3 pKa = 5.84SHH5 pKa = 6.74PARR8 pKa = 11.84RR9 pKa = 11.84SPLRR13 pKa = 11.84ALAGAAAVVVGLVLSGCTAGPDD35 pKa = 3.54TGAVTAPPEE44 pKa = 4.35VIGEE48 pKa = 4.16VPEE51 pKa = 4.29DD52 pKa = 3.42LRR54 pKa = 11.84EE55 pKa = 4.19FYY57 pKa = 10.75AQEE60 pKa = 4.27VTWEE64 pKa = 4.12EE65 pKa = 4.54CEE67 pKa = 4.47GDD69 pKa = 4.32DD70 pKa = 5.19FACATVKK77 pKa = 10.57VPLDD81 pKa = 3.45YY82 pKa = 11.03SDD84 pKa = 4.6PGRR87 pKa = 11.84ASIDD91 pKa = 2.9IAMIRR96 pKa = 11.84LDD98 pKa = 4.4APGDD102 pKa = 3.81AQGSLLVNPGGPGVSGYY119 pKa = 11.17NLVRR123 pKa = 11.84DD124 pKa = 3.72SAAFILSDD132 pKa = 4.09RR133 pKa = 11.84LRR135 pKa = 11.84QSFDD139 pKa = 3.18VVGFDD144 pKa = 3.37PRR146 pKa = 11.84GVNEE150 pKa = 4.16SSAVEE155 pKa = 4.1CLTDD159 pKa = 3.48EE160 pKa = 4.52EE161 pKa = 4.67RR162 pKa = 11.84DD163 pKa = 3.4EE164 pKa = 5.03ARR166 pKa = 11.84AEE168 pKa = 4.11TLPPDD173 pKa = 3.87ATDD176 pKa = 4.06AEE178 pKa = 4.21ALEE181 pKa = 4.19ILEE184 pKa = 4.79ADD186 pKa = 3.53SAEE189 pKa = 4.14YY190 pKa = 10.51AQLCAEE196 pKa = 4.32RR197 pKa = 11.84TGEE200 pKa = 4.09LLGHH204 pKa = 7.11IDD206 pKa = 3.58TVSAAKK212 pKa = 10.9DD213 pKa = 3.4MDD215 pKa = 3.52ILRR218 pKa = 11.84AVLGDD223 pKa = 3.5EE224 pKa = 3.91QLNYY228 pKa = 10.92LGFSYY233 pKa = 8.74GTQLGAAYY241 pKa = 10.26AGLFPGNVGRR251 pKa = 11.84FVLDD255 pKa = 3.69GAVDD259 pKa = 4.12PSLSSAEE266 pKa = 3.8ITAGQVAAFDD276 pKa = 3.67EE277 pKa = 4.39ALRR280 pKa = 11.84NYY282 pKa = 9.87VADD285 pKa = 3.99CQSQSEE291 pKa = 4.67CPLPGNVDD299 pKa = 3.18EE300 pKa = 5.38GVAVVDD306 pKa = 3.87EE307 pKa = 4.57LFEE310 pKa = 4.39SVEE313 pKa = 4.14EE314 pKa = 4.43SPMTAADD321 pKa = 3.92GRR323 pKa = 11.84PVTIGTFFQGFVLPLYY339 pKa = 10.75DD340 pKa = 3.69SASWPALTAAIDD352 pKa = 3.45QALQGDD358 pKa = 4.0PSEE361 pKa = 4.52FLRR364 pKa = 11.84FADD367 pKa = 4.44LGADD371 pKa = 3.85RR372 pKa = 11.84SPTGDD377 pKa = 3.56YY378 pKa = 8.55LTNSAAAFTAVTCLDD393 pKa = 3.85YY394 pKa = 11.23PMPADD399 pKa = 3.98PGVMAEE405 pKa = 3.97EE406 pKa = 4.59AEE408 pKa = 4.09ALEE411 pKa = 4.44ASSAVFGEE419 pKa = 4.12YY420 pKa = 10.4LSYY423 pKa = 11.15GALACKK429 pKa = 9.69NWAYY433 pKa = 10.52EE434 pKa = 4.13PVNEE438 pKa = 4.7PSPASAPGAPPMLVIGTTGDD458 pKa = 3.36PATPYY463 pKa = 9.88EE464 pKa = 4.19WSLALAEE471 pKa = 4.11QLEE474 pKa = 4.9SAVHH478 pKa = 4.68VTWEE482 pKa = 4.56GQGHH486 pKa = 5.01TAYY489 pKa = 10.14GRR491 pKa = 11.84SGEE494 pKa = 4.41CIEE497 pKa = 4.92SLVDD501 pKa = 4.77DD502 pKa = 4.49FLIDD506 pKa = 3.45GTVPEE511 pKa = 4.47MSARR515 pKa = 11.84CC516 pKa = 3.57
Molecular weight: 54.21 kDa
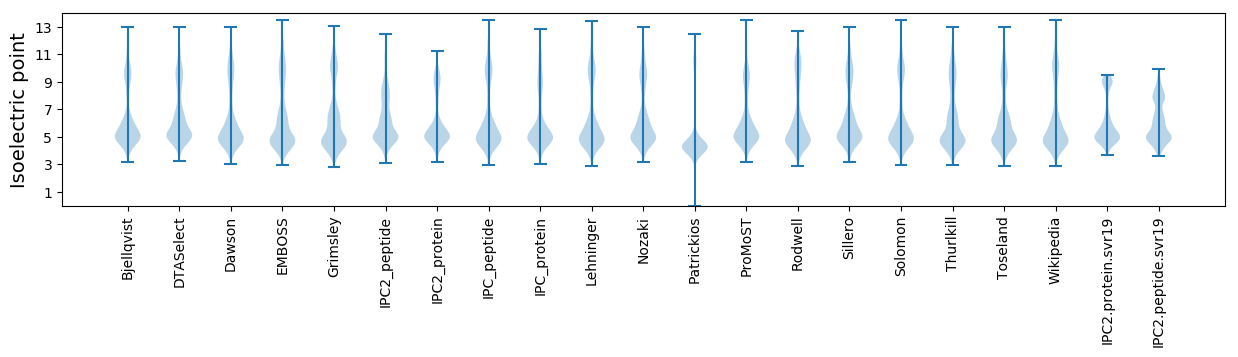
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G8GL13|A0A1G8GL13_9MICC Myo-inositol-1(Or 4)-monophosphatase OS=Arthrobacter subterraneus OX=335973 GN=SAMN04488693_104171 PE=4 SV=1
MM1 pKa = 7.4GLSNAIRR8 pKa = 11.84GAAGRR13 pKa = 11.84FTGRR17 pKa = 11.84SGGTTGRR24 pKa = 11.84TPRR27 pKa = 11.84TGGMGTGRR35 pKa = 11.84GGVSRR40 pKa = 11.84GGSGGVGGILRR51 pKa = 11.84GILRR55 pKa = 11.84RR56 pKa = 11.84RR57 pKa = 3.46
MM1 pKa = 7.4GLSNAIRR8 pKa = 11.84GAAGRR13 pKa = 11.84FTGRR17 pKa = 11.84SGGTTGRR24 pKa = 11.84TPRR27 pKa = 11.84TGGMGTGRR35 pKa = 11.84GGVSRR40 pKa = 11.84GGSGGVGGILRR51 pKa = 11.84GILRR55 pKa = 11.84RR56 pKa = 11.84RR57 pKa = 3.46
Molecular weight: 5.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1160108 |
19 |
2041 |
320.1 |
34.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.554 ± 0.056 | 0.616 ± 0.009 |
5.738 ± 0.035 | 6.044 ± 0.038 |
3.169 ± 0.025 | 8.972 ± 0.035 |
2.113 ± 0.018 | 4.39 ± 0.032 |
2.318 ± 0.028 | 10.311 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.951 ± 0.018 | 2.461 ± 0.021 |
5.444 ± 0.033 | 3.11 ± 0.02 |
6.939 ± 0.043 | 5.992 ± 0.027 |
5.946 ± 0.03 | 8.429 ± 0.038 |
1.42 ± 0.019 | 2.083 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
