
Torque teno virus (isolate Human/Japan/SANBAN/1999) (TTV)
Taxonomy: Viruses; Anelloviridae; unclassified Anelloviridae; Torque teno virus; TT virus group 3
Average proteome isoelectric point is 8.88
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

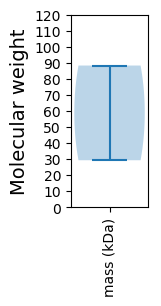
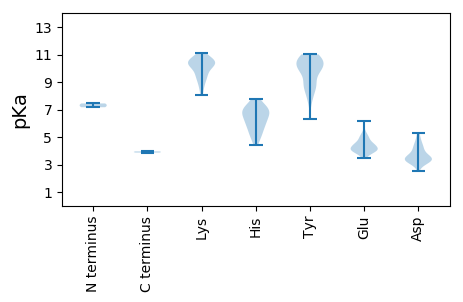
Protein with the lowest isoelectric point:
>sp|Q9WSV8|ORF23_TTVV5 Probable protein VP2 OS=Torque teno virus (isolate Human/Japan/SANBAN/1999) OX=486277 GN=ORF2/3 PE=3 SV=2
MM1 pKa = 7.44AWGWWRR7 pKa = 11.84RR8 pKa = 11.84WRR10 pKa = 11.84RR11 pKa = 11.84WPARR15 pKa = 11.84RR16 pKa = 11.84WRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84PLRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84AGRR33 pKa = 11.84PARR36 pKa = 11.84RR37 pKa = 11.84YY38 pKa = 8.56RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84TVRR45 pKa = 11.84TRR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84WGRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84YY56 pKa = 8.99RR57 pKa = 11.84RR58 pKa = 11.84GWRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84TYY65 pKa = 9.77VRR67 pKa = 11.84KK68 pKa = 9.82GRR70 pKa = 11.84HH71 pKa = 4.43RR72 pKa = 11.84KK73 pKa = 8.58KK74 pKa = 10.35KK75 pKa = 9.62KK76 pKa = 10.13RR77 pKa = 11.84LILRR81 pKa = 11.84QWQPATRR88 pKa = 11.84RR89 pKa = 11.84RR90 pKa = 11.84CTITGYY96 pKa = 10.9LPIVFCGHH104 pKa = 5.39TKK106 pKa = 10.38GNKK109 pKa = 9.43NYY111 pKa = 10.35ALHH114 pKa = 6.91SDD116 pKa = 4.09DD117 pKa = 4.72YY118 pKa = 10.39TPQGQPFGGALSTTSFSLKK137 pKa = 10.54VLFDD141 pKa = 2.86QHH143 pKa = 6.7QRR145 pKa = 11.84GLNKK149 pKa = 9.43WSFPNDD155 pKa = 3.33QLDD158 pKa = 3.49LARR161 pKa = 11.84YY162 pKa = 8.61RR163 pKa = 11.84GCKK166 pKa = 9.83FIFYY170 pKa = 7.93RR171 pKa = 11.84TKK173 pKa = 8.03QTDD176 pKa = 3.23WIGQYY181 pKa = 10.53DD182 pKa = 3.21ISEE185 pKa = 4.46PYY187 pKa = 10.63KK188 pKa = 10.32LDD190 pKa = 3.81KK191 pKa = 11.02YY192 pKa = 10.83SCPNYY197 pKa = 10.43HH198 pKa = 7.26PGNLIKK204 pKa = 10.6AKK206 pKa = 10.17HH207 pKa = 5.88KK208 pKa = 10.45FLIPSYY214 pKa = 9.03DD215 pKa = 3.43TNPRR219 pKa = 11.84GRR221 pKa = 11.84QKK223 pKa = 10.5IIVKK227 pKa = 10.06IPPPDD232 pKa = 3.44LFVDD236 pKa = 3.59KK237 pKa = 10.35WYY239 pKa = 8.82TQEE242 pKa = 4.91DD243 pKa = 4.53LCSVNLVSLAVSAASFLHH261 pKa = 6.42PFGSPQTDD269 pKa = 3.38NPCYY273 pKa = 9.34TFQVLKK279 pKa = 10.39EE280 pKa = 4.24FYY282 pKa = 8.67YY283 pKa = 10.4QAIGFSATDD292 pKa = 3.17EE293 pKa = 4.13SRR295 pKa = 11.84NYY297 pKa = 9.48VFNVLYY303 pKa = 10.44EE304 pKa = 4.18EE305 pKa = 4.04NSYY308 pKa = 9.95WEE310 pKa = 4.4SNITPFYY317 pKa = 10.0VINVKK322 pKa = 10.39KK323 pKa = 10.83GSNTXDD329 pKa = 3.46YY330 pKa = 7.7MSPXISDD337 pKa = 3.07SHH339 pKa = 6.55FRR341 pKa = 11.84NKK343 pKa = 9.96VNTNYY348 pKa = 10.32NWYY351 pKa = 8.09TYY353 pKa = 8.8NAKK356 pKa = 9.05SHH358 pKa = 6.75KK359 pKa = 10.18NDD361 pKa = 3.0LHH363 pKa = 5.16XLRR366 pKa = 11.84RR367 pKa = 11.84AYY369 pKa = 9.99FKK371 pKa = 10.84QLTTEE376 pKa = 5.26GPQQTSSEE384 pKa = 4.13KK385 pKa = 10.72GYY387 pKa = 10.77ASQWTTPTTDD397 pKa = 2.49AYY399 pKa = 9.96EE400 pKa = 4.0YY401 pKa = 9.93HH402 pKa = 7.78LGMFSTIFLAPDD414 pKa = 3.23RR415 pKa = 11.84PVPRR419 pKa = 11.84FPCAYY424 pKa = 10.13QDD426 pKa = 3.4VTYY429 pKa = 10.93NPLMDD434 pKa = 4.37KK435 pKa = 11.08GVGNHH440 pKa = 5.86VWFQYY445 pKa = 7.57NTKK448 pKa = 10.57ADD450 pKa = 3.56TQLIVTGGSCKK461 pKa = 10.55AHH463 pKa = 7.31IEE465 pKa = 5.03DD466 pKa = 3.86IPLWAAFYY474 pKa = 10.09GYY476 pKa = 10.81SDD478 pKa = 4.68FIEE481 pKa = 4.89SEE483 pKa = 4.01LGPFVDD489 pKa = 4.9AEE491 pKa = 4.69TVGLICVICPYY502 pKa = 9.55TKK504 pKa = 10.07PPMYY508 pKa = 10.75NKK510 pKa = 8.53TNPMMGYY517 pKa = 9.93VFYY520 pKa = 10.94DD521 pKa = 3.3RR522 pKa = 11.84NFGDD526 pKa = 4.44GKK528 pKa = 10.1WIDD531 pKa = 3.65GRR533 pKa = 11.84GKK535 pKa = 9.76IEE537 pKa = 4.95PYY539 pKa = 8.88WQVRR543 pKa = 11.84WRR545 pKa = 11.84PEE547 pKa = 3.47MLFQEE552 pKa = 4.6TVMADD557 pKa = 3.2IVQTGPFSYY566 pKa = 10.56KK567 pKa = 10.63DD568 pKa = 3.45EE569 pKa = 4.92LKK571 pKa = 11.1NSTLVAKK578 pKa = 10.44YY579 pKa = 10.31KK580 pKa = 10.65FYY582 pKa = 9.95FTWGGNMMFQQTIKK596 pKa = 10.75NPCKK600 pKa = 9.64TDD602 pKa = 3.15GRR604 pKa = 11.84PTDD607 pKa = 3.43SDD609 pKa = 3.35RR610 pKa = 11.84HH611 pKa = 5.0PRR613 pKa = 11.84GIQVADD619 pKa = 3.96PEE621 pKa = 4.54QMGPRR626 pKa = 11.84WVFHH630 pKa = 5.99SFDD633 pKa = 2.91WRR635 pKa = 11.84RR636 pKa = 11.84GYY638 pKa = 10.88LSEE641 pKa = 4.14GAIKK645 pKa = 10.39RR646 pKa = 11.84LHH648 pKa = 6.42EE649 pKa = 4.53KK650 pKa = 10.18PLDD653 pKa = 3.56YY654 pKa = 10.75EE655 pKa = 5.01SYY657 pKa = 8.49FTQPKK662 pKa = 9.16RR663 pKa = 11.84PRR665 pKa = 11.84IFPPTEE671 pKa = 3.54AAEE674 pKa = 4.62GEE676 pKa = 4.1FRR678 pKa = 11.84EE679 pKa = 4.5PEE681 pKa = 3.87KK682 pKa = 10.94GSYY685 pKa = 9.93SEE687 pKa = 4.54EE688 pKa = 3.9EE689 pKa = 3.94RR690 pKa = 11.84SQASAEE696 pKa = 4.09EE697 pKa = 4.14QATEE701 pKa = 4.07EE702 pKa = 4.21TVLLLKK708 pKa = 10.53RR709 pKa = 11.84RR710 pKa = 11.84LRR712 pKa = 11.84EE713 pKa = 3.8QRR715 pKa = 11.84KK716 pKa = 9.48LEE718 pKa = 3.91QQLQFLTRR726 pKa = 11.84EE727 pKa = 4.07MFKK730 pKa = 9.1TQAGLHH736 pKa = 5.98INPMLLSQRR745 pKa = 3.87
MM1 pKa = 7.44AWGWWRR7 pKa = 11.84RR8 pKa = 11.84WRR10 pKa = 11.84RR11 pKa = 11.84WPARR15 pKa = 11.84RR16 pKa = 11.84WRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84PLRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84AGRR33 pKa = 11.84PARR36 pKa = 11.84RR37 pKa = 11.84YY38 pKa = 8.56RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84TVRR45 pKa = 11.84TRR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84WGRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84YY56 pKa = 8.99RR57 pKa = 11.84RR58 pKa = 11.84GWRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84TYY65 pKa = 9.77VRR67 pKa = 11.84KK68 pKa = 9.82GRR70 pKa = 11.84HH71 pKa = 4.43RR72 pKa = 11.84KK73 pKa = 8.58KK74 pKa = 10.35KK75 pKa = 9.62KK76 pKa = 10.13RR77 pKa = 11.84LILRR81 pKa = 11.84QWQPATRR88 pKa = 11.84RR89 pKa = 11.84RR90 pKa = 11.84CTITGYY96 pKa = 10.9LPIVFCGHH104 pKa = 5.39TKK106 pKa = 10.38GNKK109 pKa = 9.43NYY111 pKa = 10.35ALHH114 pKa = 6.91SDD116 pKa = 4.09DD117 pKa = 4.72YY118 pKa = 10.39TPQGQPFGGALSTTSFSLKK137 pKa = 10.54VLFDD141 pKa = 2.86QHH143 pKa = 6.7QRR145 pKa = 11.84GLNKK149 pKa = 9.43WSFPNDD155 pKa = 3.33QLDD158 pKa = 3.49LARR161 pKa = 11.84YY162 pKa = 8.61RR163 pKa = 11.84GCKK166 pKa = 9.83FIFYY170 pKa = 7.93RR171 pKa = 11.84TKK173 pKa = 8.03QTDD176 pKa = 3.23WIGQYY181 pKa = 10.53DD182 pKa = 3.21ISEE185 pKa = 4.46PYY187 pKa = 10.63KK188 pKa = 10.32LDD190 pKa = 3.81KK191 pKa = 11.02YY192 pKa = 10.83SCPNYY197 pKa = 10.43HH198 pKa = 7.26PGNLIKK204 pKa = 10.6AKK206 pKa = 10.17HH207 pKa = 5.88KK208 pKa = 10.45FLIPSYY214 pKa = 9.03DD215 pKa = 3.43TNPRR219 pKa = 11.84GRR221 pKa = 11.84QKK223 pKa = 10.5IIVKK227 pKa = 10.06IPPPDD232 pKa = 3.44LFVDD236 pKa = 3.59KK237 pKa = 10.35WYY239 pKa = 8.82TQEE242 pKa = 4.91DD243 pKa = 4.53LCSVNLVSLAVSAASFLHH261 pKa = 6.42PFGSPQTDD269 pKa = 3.38NPCYY273 pKa = 9.34TFQVLKK279 pKa = 10.39EE280 pKa = 4.24FYY282 pKa = 8.67YY283 pKa = 10.4QAIGFSATDD292 pKa = 3.17EE293 pKa = 4.13SRR295 pKa = 11.84NYY297 pKa = 9.48VFNVLYY303 pKa = 10.44EE304 pKa = 4.18EE305 pKa = 4.04NSYY308 pKa = 9.95WEE310 pKa = 4.4SNITPFYY317 pKa = 10.0VINVKK322 pKa = 10.39KK323 pKa = 10.83GSNTXDD329 pKa = 3.46YY330 pKa = 7.7MSPXISDD337 pKa = 3.07SHH339 pKa = 6.55FRR341 pKa = 11.84NKK343 pKa = 9.96VNTNYY348 pKa = 10.32NWYY351 pKa = 8.09TYY353 pKa = 8.8NAKK356 pKa = 9.05SHH358 pKa = 6.75KK359 pKa = 10.18NDD361 pKa = 3.0LHH363 pKa = 5.16XLRR366 pKa = 11.84RR367 pKa = 11.84AYY369 pKa = 9.99FKK371 pKa = 10.84QLTTEE376 pKa = 5.26GPQQTSSEE384 pKa = 4.13KK385 pKa = 10.72GYY387 pKa = 10.77ASQWTTPTTDD397 pKa = 2.49AYY399 pKa = 9.96EE400 pKa = 4.0YY401 pKa = 9.93HH402 pKa = 7.78LGMFSTIFLAPDD414 pKa = 3.23RR415 pKa = 11.84PVPRR419 pKa = 11.84FPCAYY424 pKa = 10.13QDD426 pKa = 3.4VTYY429 pKa = 10.93NPLMDD434 pKa = 4.37KK435 pKa = 11.08GVGNHH440 pKa = 5.86VWFQYY445 pKa = 7.57NTKK448 pKa = 10.57ADD450 pKa = 3.56TQLIVTGGSCKK461 pKa = 10.55AHH463 pKa = 7.31IEE465 pKa = 5.03DD466 pKa = 3.86IPLWAAFYY474 pKa = 10.09GYY476 pKa = 10.81SDD478 pKa = 4.68FIEE481 pKa = 4.89SEE483 pKa = 4.01LGPFVDD489 pKa = 4.9AEE491 pKa = 4.69TVGLICVICPYY502 pKa = 9.55TKK504 pKa = 10.07PPMYY508 pKa = 10.75NKK510 pKa = 8.53TNPMMGYY517 pKa = 9.93VFYY520 pKa = 10.94DD521 pKa = 3.3RR522 pKa = 11.84NFGDD526 pKa = 4.44GKK528 pKa = 10.1WIDD531 pKa = 3.65GRR533 pKa = 11.84GKK535 pKa = 9.76IEE537 pKa = 4.95PYY539 pKa = 8.88WQVRR543 pKa = 11.84WRR545 pKa = 11.84PEE547 pKa = 3.47MLFQEE552 pKa = 4.6TVMADD557 pKa = 3.2IVQTGPFSYY566 pKa = 10.56KK567 pKa = 10.63DD568 pKa = 3.45EE569 pKa = 4.92LKK571 pKa = 11.1NSTLVAKK578 pKa = 10.44YY579 pKa = 10.31KK580 pKa = 10.65FYY582 pKa = 9.95FTWGGNMMFQQTIKK596 pKa = 10.75NPCKK600 pKa = 9.64TDD602 pKa = 3.15GRR604 pKa = 11.84PTDD607 pKa = 3.43SDD609 pKa = 3.35RR610 pKa = 11.84HH611 pKa = 5.0PRR613 pKa = 11.84GIQVADD619 pKa = 3.96PEE621 pKa = 4.54QMGPRR626 pKa = 11.84WVFHH630 pKa = 5.99SFDD633 pKa = 2.91WRR635 pKa = 11.84RR636 pKa = 11.84GYY638 pKa = 10.88LSEE641 pKa = 4.14GAIKK645 pKa = 10.39RR646 pKa = 11.84LHH648 pKa = 6.42EE649 pKa = 4.53KK650 pKa = 10.18PLDD653 pKa = 3.56YY654 pKa = 10.75EE655 pKa = 5.01SYY657 pKa = 8.49FTQPKK662 pKa = 9.16RR663 pKa = 11.84PRR665 pKa = 11.84IFPPTEE671 pKa = 3.54AAEE674 pKa = 4.62GEE676 pKa = 4.1FRR678 pKa = 11.84EE679 pKa = 4.5PEE681 pKa = 3.87KK682 pKa = 10.94GSYY685 pKa = 9.93SEE687 pKa = 4.54EE688 pKa = 3.9EE689 pKa = 3.94RR690 pKa = 11.84SQASAEE696 pKa = 4.09EE697 pKa = 4.14QATEE701 pKa = 4.07EE702 pKa = 4.21TVLLLKK708 pKa = 10.53RR709 pKa = 11.84RR710 pKa = 11.84LRR712 pKa = 11.84EE713 pKa = 3.8QRR715 pKa = 11.84KK716 pKa = 9.48LEE718 pKa = 3.91QQLQFLTRR726 pKa = 11.84EE727 pKa = 4.07MFKK730 pKa = 9.1TQAGLHH736 pKa = 5.98INPMLLSQRR745 pKa = 3.87
Molecular weight: 88.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9WSV8|ORF23_TTVV5 Probable protein VP2 OS=Torque teno virus (isolate Human/Japan/SANBAN/1999) OX=486277 GN=ORF2/3 PE=3 SV=2
MM1 pKa = 7.17SWRR4 pKa = 11.84PPVHH8 pKa = 6.97DD9 pKa = 4.22APGIEE14 pKa = 4.18RR15 pKa = 11.84NWYY18 pKa = 6.33EE19 pKa = 3.73ACFRR23 pKa = 11.84AHH25 pKa = 7.17AGSCGCGNFIAHH37 pKa = 6.83INLLAGRR44 pKa = 11.84YY45 pKa = 8.5GFTGGPPPPGGPPPGTPQVRR65 pKa = 11.84ASRR68 pKa = 11.84NSPAAPQQPPALPWHH83 pKa = 6.77GDD85 pKa = 3.2GGEE88 pKa = 4.37GGAAGPPGAGGDD100 pKa = 3.76AAADD104 pKa = 3.26AHH106 pKa = 7.23LGDD109 pKa = 5.07EE110 pKa = 4.49EE111 pKa = 5.42LADD114 pKa = 4.33LLDD117 pKa = 5.27AIEE120 pKa = 6.17DD121 pKa = 3.9DD122 pKa = 4.22AQCSNRR128 pKa = 11.84RR129 pKa = 11.84SKK131 pKa = 9.6TRR133 pKa = 11.84ARR135 pKa = 11.84RR136 pKa = 11.84TDD138 pKa = 3.64GPPTPIDD145 pKa = 3.35TLEE148 pKa = 4.0EE149 pKa = 4.41YY150 pKa = 10.12KK151 pKa = 10.33WRR153 pKa = 11.84TRR155 pKa = 11.84NKK157 pKa = 8.83WDD159 pKa = 3.55PAGCSTPLTGEE170 pKa = 3.93GAILARR176 pKa = 11.84EE177 pKa = 4.27LSNACTRR184 pKa = 11.84NLSTMKK190 pKa = 10.63AILHH194 pKa = 5.21NQKK197 pKa = 10.5DD198 pKa = 4.2LEE200 pKa = 4.4SFLQQRR206 pKa = 11.84QQRR209 pKa = 11.84EE210 pKa = 4.13SSEE213 pKa = 4.41SPKK216 pKa = 10.15KK217 pKa = 10.79AHH219 pKa = 5.95IQRR222 pKa = 11.84KK223 pKa = 9.1KK224 pKa = 10.14GRR226 pKa = 11.84KK227 pKa = 8.14PLQKK231 pKa = 9.3SRR233 pKa = 11.84RR234 pKa = 11.84RR235 pKa = 11.84RR236 pKa = 11.84RR237 pKa = 11.84QYY239 pKa = 11.01SSSSDD244 pKa = 3.41DD245 pKa = 3.87SEE247 pKa = 5.16SSGSSSSSSNSSPEE261 pKa = 3.73KK262 pKa = 8.75CSKK265 pKa = 10.13RR266 pKa = 11.84KK267 pKa = 9.46RR268 pKa = 11.84VSTT271 pKa = 3.96
MM1 pKa = 7.17SWRR4 pKa = 11.84PPVHH8 pKa = 6.97DD9 pKa = 4.22APGIEE14 pKa = 4.18RR15 pKa = 11.84NWYY18 pKa = 6.33EE19 pKa = 3.73ACFRR23 pKa = 11.84AHH25 pKa = 7.17AGSCGCGNFIAHH37 pKa = 6.83INLLAGRR44 pKa = 11.84YY45 pKa = 8.5GFTGGPPPPGGPPPGTPQVRR65 pKa = 11.84ASRR68 pKa = 11.84NSPAAPQQPPALPWHH83 pKa = 6.77GDD85 pKa = 3.2GGEE88 pKa = 4.37GGAAGPPGAGGDD100 pKa = 3.76AAADD104 pKa = 3.26AHH106 pKa = 7.23LGDD109 pKa = 5.07EE110 pKa = 4.49EE111 pKa = 5.42LADD114 pKa = 4.33LLDD117 pKa = 5.27AIEE120 pKa = 6.17DD121 pKa = 3.9DD122 pKa = 4.22AQCSNRR128 pKa = 11.84RR129 pKa = 11.84SKK131 pKa = 9.6TRR133 pKa = 11.84ARR135 pKa = 11.84RR136 pKa = 11.84TDD138 pKa = 3.64GPPTPIDD145 pKa = 3.35TLEE148 pKa = 4.0EE149 pKa = 4.41YY150 pKa = 10.12KK151 pKa = 10.33WRR153 pKa = 11.84TRR155 pKa = 11.84NKK157 pKa = 8.83WDD159 pKa = 3.55PAGCSTPLTGEE170 pKa = 3.93GAILARR176 pKa = 11.84EE177 pKa = 4.27LSNACTRR184 pKa = 11.84NLSTMKK190 pKa = 10.63AILHH194 pKa = 5.21NQKK197 pKa = 10.5DD198 pKa = 4.2LEE200 pKa = 4.4SFLQQRR206 pKa = 11.84QQRR209 pKa = 11.84EE210 pKa = 4.13SSEE213 pKa = 4.41SPKK216 pKa = 10.15KK217 pKa = 10.79AHH219 pKa = 5.95IQRR222 pKa = 11.84KK223 pKa = 9.1KK224 pKa = 10.14GRR226 pKa = 11.84KK227 pKa = 8.14PLQKK231 pKa = 9.3SRR233 pKa = 11.84RR234 pKa = 11.84RR235 pKa = 11.84RR236 pKa = 11.84RR237 pKa = 11.84QYY239 pKa = 11.01SSSSDD244 pKa = 3.41DD245 pKa = 3.87SEE247 pKa = 5.16SSGSSSSSSNSSPEE261 pKa = 3.73KK262 pKa = 8.75CSKK265 pKa = 10.13RR266 pKa = 11.84KK267 pKa = 9.46RR268 pKa = 11.84VSTT271 pKa = 3.96
Molecular weight: 29.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1016 |
271 |
745 |
508.0 |
58.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.102 ± 1.99 | 1.772 ± 0.418 |
5.02 ± 0.266 | 5.118 ± 0.215 |
4.232 ± 1.421 | 7.087 ± 1.483 |
2.362 ± 0.114 | 3.74 ± 0.406 |
6.004 ± 0.432 | 6.201 ± 0.153 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.575 ± 0.431 | 4.035 ± 0.178 |
7.382 ± 1.14 | 4.823 ± 0.204 |
10.236 ± 0.331 | 6.988 ± 2.294 |
6.004 ± 0.812 | 3.346 ± 1.154 |
2.756 ± 0.47 | 4.921 ± 1.776 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
