
Bacilladnaviridae sp.
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Baphyvirales; Bacilladnaviridae; unclassified Bacilladnaviridae
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
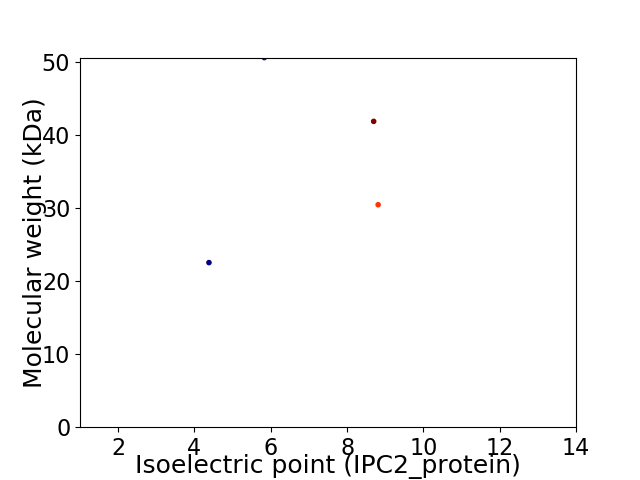
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
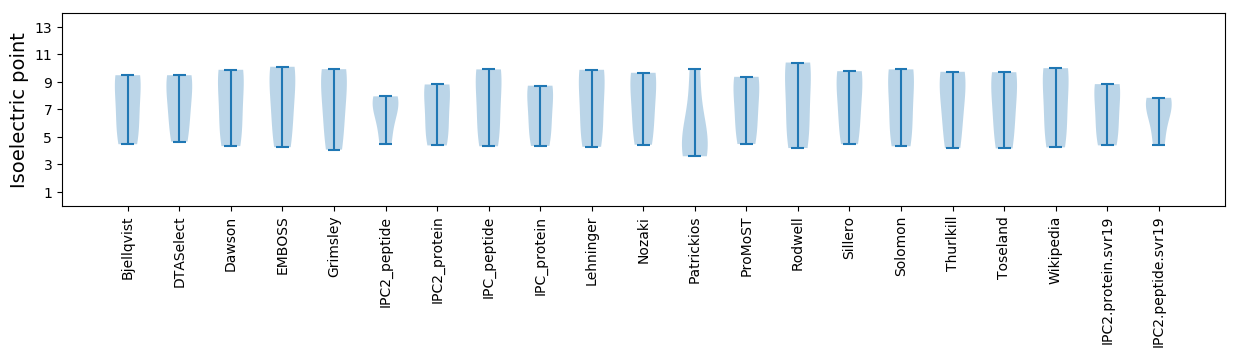
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345MP94|A0A345MP94_9VIRU Uncharacterized protein OS=Bacilladnaviridae sp. OX=2202647 PE=4 SV=1
MM1 pKa = 7.5SIKK4 pKa = 10.39PIMRR8 pKa = 11.84SVSVPTVLPDD18 pKa = 3.12SQMYY22 pKa = 9.6FLHH25 pKa = 6.97RR26 pKa = 11.84NAGTEE31 pKa = 3.78LLDD34 pKa = 3.38YY35 pKa = 11.05HH36 pKa = 7.16FDD38 pKa = 3.65VLSGLIYY45 pKa = 10.83LNARR49 pKa = 11.84GHH51 pKa = 5.39RR52 pKa = 11.84TVHH55 pKa = 5.41YY56 pKa = 10.8VRR58 pKa = 11.84DD59 pKa = 4.18LIKK62 pKa = 10.91AIDD65 pKa = 3.77DD66 pKa = 3.13WHH68 pKa = 6.38YY69 pKa = 10.94RR70 pKa = 11.84ALVDD74 pKa = 5.19DD75 pKa = 4.54RR76 pKa = 11.84EE77 pKa = 4.26KK78 pKa = 11.24EE79 pKa = 3.91QLQQLILVDD88 pKa = 4.24LMTNEE93 pKa = 4.48EE94 pKa = 4.48LYY96 pKa = 10.84KK97 pKa = 10.92DD98 pKa = 3.56AVIWDD103 pKa = 4.04GTRR106 pKa = 11.84KK107 pKa = 9.71RR108 pKa = 11.84YY109 pKa = 9.68YY110 pKa = 10.9VIDD113 pKa = 3.03AWIRR117 pKa = 11.84NHH119 pKa = 5.1STCFEE124 pKa = 4.03FTGPIDD130 pKa = 3.57PANIKK135 pKa = 10.23VEE137 pKa = 4.23EE138 pKa = 4.26EE139 pKa = 4.17EE140 pKa = 4.42PTNLDD145 pKa = 3.34DD146 pKa = 4.08TFSLGTITTFTMDD159 pKa = 4.13EE160 pKa = 4.42SPLGLVGGAIEE171 pKa = 5.13FDD173 pKa = 3.38WEE175 pKa = 4.24LDD177 pKa = 3.73SNGYY181 pKa = 9.32LLNDD185 pKa = 3.78FVVTVDD191 pKa = 4.8DD192 pKa = 4.31PLII195 pKa = 4.45
MM1 pKa = 7.5SIKK4 pKa = 10.39PIMRR8 pKa = 11.84SVSVPTVLPDD18 pKa = 3.12SQMYY22 pKa = 9.6FLHH25 pKa = 6.97RR26 pKa = 11.84NAGTEE31 pKa = 3.78LLDD34 pKa = 3.38YY35 pKa = 11.05HH36 pKa = 7.16FDD38 pKa = 3.65VLSGLIYY45 pKa = 10.83LNARR49 pKa = 11.84GHH51 pKa = 5.39RR52 pKa = 11.84TVHH55 pKa = 5.41YY56 pKa = 10.8VRR58 pKa = 11.84DD59 pKa = 4.18LIKK62 pKa = 10.91AIDD65 pKa = 3.77DD66 pKa = 3.13WHH68 pKa = 6.38YY69 pKa = 10.94RR70 pKa = 11.84ALVDD74 pKa = 5.19DD75 pKa = 4.54RR76 pKa = 11.84EE77 pKa = 4.26KK78 pKa = 11.24EE79 pKa = 3.91QLQQLILVDD88 pKa = 4.24LMTNEE93 pKa = 4.48EE94 pKa = 4.48LYY96 pKa = 10.84KK97 pKa = 10.92DD98 pKa = 3.56AVIWDD103 pKa = 4.04GTRR106 pKa = 11.84KK107 pKa = 9.71RR108 pKa = 11.84YY109 pKa = 9.68YY110 pKa = 10.9VIDD113 pKa = 3.03AWIRR117 pKa = 11.84NHH119 pKa = 5.1STCFEE124 pKa = 4.03FTGPIDD130 pKa = 3.57PANIKK135 pKa = 10.23VEE137 pKa = 4.23EE138 pKa = 4.26EE139 pKa = 4.17EE140 pKa = 4.42PTNLDD145 pKa = 3.34DD146 pKa = 4.08TFSLGTITTFTMDD159 pKa = 4.13EE160 pKa = 4.42SPLGLVGGAIEE171 pKa = 5.13FDD173 pKa = 3.38WEE175 pKa = 4.24LDD177 pKa = 3.73SNGYY181 pKa = 9.32LLNDD185 pKa = 3.78FVVTVDD191 pKa = 4.8DD192 pKa = 4.31PLII195 pKa = 4.45
Molecular weight: 22.52 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345MP95|A0A345MP95_9VIRU Putative esterase/lipase OS=Bacilladnaviridae sp. OX=2202647 PE=4 SV=1
MM1 pKa = 7.43VKK3 pKa = 10.19RR4 pKa = 11.84KK5 pKa = 8.58RR6 pKa = 11.84TIGGPRR12 pKa = 11.84RR13 pKa = 11.84KK14 pKa = 8.67STRR17 pKa = 11.84QTKK20 pKa = 9.97KK21 pKa = 7.55RR22 pKa = 11.84TKK24 pKa = 7.3WTSRR28 pKa = 11.84SYY30 pKa = 11.01TPAPKK35 pKa = 9.62PKK37 pKa = 9.48KK38 pKa = 9.87RR39 pKa = 11.84KK40 pKa = 9.11ASKK43 pKa = 9.47PRR45 pKa = 11.84YY46 pKa = 8.31AAQHH50 pKa = 5.3HH51 pKa = 7.3AYY53 pKa = 8.87MKK55 pKa = 10.98SPFSKK60 pKa = 10.54NASKK64 pKa = 10.53QPKK67 pKa = 9.23IPDD70 pKa = 3.6GAFTTSMGRR79 pKa = 11.84AFEE82 pKa = 4.53TIAEE86 pKa = 4.19IQNAVGNEE94 pKa = 4.12TIDD97 pKa = 3.1IVLFPGFGIGAAIQNTTAAATMDD120 pKa = 4.04GVQPIALDD128 pKa = 4.27HH129 pKa = 5.47STNWLGSADD138 pKa = 3.6TSKK141 pKa = 11.26SVDD144 pKa = 3.55NEE146 pKa = 4.15SEE148 pKa = 3.87IARR151 pKa = 11.84WRR153 pKa = 11.84LVAQSLQLLLVNSAEE168 pKa = 4.47EE169 pKa = 3.91NNGWYY174 pKa = 6.47EE175 pKa = 4.14TCRR178 pKa = 11.84FTPAQFMADD187 pKa = 3.27YY188 pKa = 10.59VVRR191 pKa = 11.84NTDD194 pKa = 2.95GNNIEE199 pKa = 4.45NLTNAALCPHH209 pKa = 6.47NKK211 pKa = 9.47FFTDD215 pKa = 4.6HH216 pKa = 5.75ITDD219 pKa = 3.27MVMTNQRR226 pKa = 11.84GYY228 pKa = 11.22CLGKK232 pKa = 9.12LTDD235 pKa = 3.85LGKK238 pKa = 10.58KK239 pKa = 9.55EE240 pKa = 4.06FLLHH244 pKa = 6.57NISPKK249 pKa = 10.52DD250 pKa = 3.47DD251 pKa = 4.88GIINMQEE258 pKa = 3.9TYY260 pKa = 11.23NNSFANGTYY269 pKa = 10.37DD270 pKa = 5.51AINKK274 pKa = 7.44LHH276 pKa = 6.55SWTPDD281 pKa = 2.54HH282 pKa = 7.45DD283 pKa = 4.16AQIMMRR289 pKa = 11.84LLDD292 pKa = 3.76QGTSLNSHH300 pKa = 6.36DD301 pKa = 4.41MILIRR306 pKa = 11.84LHH308 pKa = 6.33CRR310 pKa = 11.84SNTGTSPNTGSKK322 pKa = 10.21FVLRR326 pKa = 11.84LGAGHH331 pKa = 5.76EE332 pKa = 4.22VEE334 pKa = 4.61YY335 pKa = 10.4TPKK338 pKa = 10.68SPLHH342 pKa = 5.56TFMTEE347 pKa = 3.66SPTVTGTNKK356 pKa = 10.46LKK358 pKa = 10.79DD359 pKa = 3.74DD360 pKa = 4.77ANNKK364 pKa = 8.67PDD366 pKa = 4.04AVTSAPFTLGGGG378 pKa = 3.64
MM1 pKa = 7.43VKK3 pKa = 10.19RR4 pKa = 11.84KK5 pKa = 8.58RR6 pKa = 11.84TIGGPRR12 pKa = 11.84RR13 pKa = 11.84KK14 pKa = 8.67STRR17 pKa = 11.84QTKK20 pKa = 9.97KK21 pKa = 7.55RR22 pKa = 11.84TKK24 pKa = 7.3WTSRR28 pKa = 11.84SYY30 pKa = 11.01TPAPKK35 pKa = 9.62PKK37 pKa = 9.48KK38 pKa = 9.87RR39 pKa = 11.84KK40 pKa = 9.11ASKK43 pKa = 9.47PRR45 pKa = 11.84YY46 pKa = 8.31AAQHH50 pKa = 5.3HH51 pKa = 7.3AYY53 pKa = 8.87MKK55 pKa = 10.98SPFSKK60 pKa = 10.54NASKK64 pKa = 10.53QPKK67 pKa = 9.23IPDD70 pKa = 3.6GAFTTSMGRR79 pKa = 11.84AFEE82 pKa = 4.53TIAEE86 pKa = 4.19IQNAVGNEE94 pKa = 4.12TIDD97 pKa = 3.1IVLFPGFGIGAAIQNTTAAATMDD120 pKa = 4.04GVQPIALDD128 pKa = 4.27HH129 pKa = 5.47STNWLGSADD138 pKa = 3.6TSKK141 pKa = 11.26SVDD144 pKa = 3.55NEE146 pKa = 4.15SEE148 pKa = 3.87IARR151 pKa = 11.84WRR153 pKa = 11.84LVAQSLQLLLVNSAEE168 pKa = 4.47EE169 pKa = 3.91NNGWYY174 pKa = 6.47EE175 pKa = 4.14TCRR178 pKa = 11.84FTPAQFMADD187 pKa = 3.27YY188 pKa = 10.59VVRR191 pKa = 11.84NTDD194 pKa = 2.95GNNIEE199 pKa = 4.45NLTNAALCPHH209 pKa = 6.47NKK211 pKa = 9.47FFTDD215 pKa = 4.6HH216 pKa = 5.75ITDD219 pKa = 3.27MVMTNQRR226 pKa = 11.84GYY228 pKa = 11.22CLGKK232 pKa = 9.12LTDD235 pKa = 3.85LGKK238 pKa = 10.58KK239 pKa = 9.55EE240 pKa = 4.06FLLHH244 pKa = 6.57NISPKK249 pKa = 10.52DD250 pKa = 3.47DD251 pKa = 4.88GIINMQEE258 pKa = 3.9TYY260 pKa = 11.23NNSFANGTYY269 pKa = 10.37DD270 pKa = 5.51AINKK274 pKa = 7.44LHH276 pKa = 6.55SWTPDD281 pKa = 2.54HH282 pKa = 7.45DD283 pKa = 4.16AQIMMRR289 pKa = 11.84LLDD292 pKa = 3.76QGTSLNSHH300 pKa = 6.36DD301 pKa = 4.41MILIRR306 pKa = 11.84LHH308 pKa = 6.33CRR310 pKa = 11.84SNTGTSPNTGSKK322 pKa = 10.21FVLRR326 pKa = 11.84LGAGHH331 pKa = 5.76EE332 pKa = 4.22VEE334 pKa = 4.61YY335 pKa = 10.4TPKK338 pKa = 10.68SPLHH342 pKa = 5.56TFMTEE347 pKa = 3.66SPTVTGTNKK356 pKa = 10.46LKK358 pKa = 10.79DD359 pKa = 3.74DD360 pKa = 4.77ANNKK364 pKa = 8.67PDD366 pKa = 4.04AVTSAPFTLGGGG378 pKa = 3.64
Molecular weight: 41.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1293 |
195 |
440 |
323.3 |
36.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.347 ± 0.871 | 0.773 ± 0.231 |
7.27 ± 0.849 | 4.872 ± 0.501 |
3.79 ± 0.444 | 6.883 ± 0.656 |
3.79 ± 0.393 | 4.872 ± 0.485 |
6.265 ± 0.754 | 7.193 ± 0.928 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.398 ± 0.349 | 5.027 ± 0.926 |
4.95 ± 0.671 | 3.48 ± 0.47 |
5.336 ± 0.15 | 6.574 ± 0.386 |
7.502 ± 0.978 | 6.342 ± 1.024 |
2.243 ± 0.409 | 3.094 ± 0.403 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
