
Mesonia sp. HuA40
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Mesonia; unclassified Mesonia
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
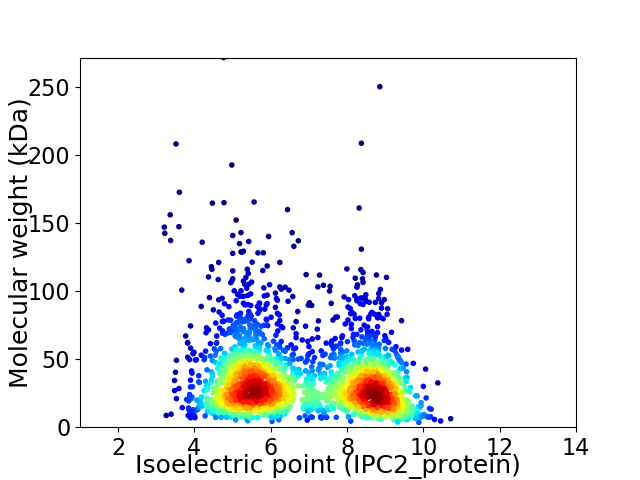
Virtual 2D-PAGE plot for 2381 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C8L413|A0A5C8L413_9FLAO DUF3810 domain-containing protein OS=Mesonia sp. HuA40 OX=2602761 GN=FT993_10855 PE=4 SV=1
MM1 pKa = 7.48KK2 pKa = 10.33KK3 pKa = 9.43ILPFILCNLLALASFAQIGINTDD26 pKa = 3.41SPSPAAALEE35 pKa = 4.33IKK37 pKa = 10.49SSYY40 pKa = 9.4NNKK43 pKa = 10.2NYY45 pKa = 10.69GGLLIPTVDD54 pKa = 2.82ATEE57 pKa = 4.02RR58 pKa = 11.84DD59 pKa = 4.02KK60 pKa = 11.4ISDD63 pKa = 3.58KK64 pKa = 10.92ATTADD69 pKa = 3.53GGMLIYY75 pKa = 10.42YY76 pKa = 9.85VEE78 pKa = 4.53GNTRR82 pKa = 11.84CLQIYY87 pKa = 10.28DD88 pKa = 4.95GINGSWADD96 pKa = 4.03VYY98 pKa = 11.46CMPTNYY104 pKa = 10.42APIASNVFISGIIEE118 pKa = 3.99DD119 pKa = 4.35TQILTANYY127 pKa = 7.87TYY129 pKa = 11.45SDD131 pKa = 3.94IEE133 pKa = 4.06NDD135 pKa = 3.7AEE137 pKa = 4.4GTTTFEE143 pKa = 3.97WFIADD148 pKa = 4.2DD149 pKa = 4.02AQGTNTTSLGVTTTGTYY166 pKa = 9.04TLQTADD172 pKa = 3.02VGKK175 pKa = 10.42FIFVRR180 pKa = 11.84VTPVASAGEE189 pKa = 4.54FIGSPVDD196 pKa = 3.48SPLRR200 pKa = 11.84GPIEE204 pKa = 4.02AAGGVASDD212 pKa = 4.37LFISEE217 pKa = 4.44YY218 pKa = 11.2VEE220 pKa = 3.96GSSYY224 pKa = 11.62NKK226 pKa = 9.94IIEE229 pKa = 4.18IANFTGQDD237 pKa = 3.47VNLSEE242 pKa = 3.87YY243 pKa = 9.94HH244 pKa = 6.49LRR246 pKa = 11.84IFRR249 pKa = 11.84QTGNLTGPISLTSNTLIDD267 pKa = 3.7GDD269 pKa = 4.12VFVIANTANVLATGEE284 pKa = 4.33VDD286 pKa = 3.49TQTGSLNFNGDD297 pKa = 3.78DD298 pKa = 4.07SVLLYY303 pKa = 8.39YY304 pKa = 10.49QPLGGTEE311 pKa = 4.02TLIDD315 pKa = 4.01IIGPDD320 pKa = 3.59SLVNPPSNNPNFAKK334 pKa = 10.72DD335 pKa = 3.12EE336 pKa = 4.17TLRR339 pKa = 11.84RR340 pKa = 11.84KK341 pKa = 9.86PGLGPSTTYY350 pKa = 11.05DD351 pKa = 3.45SNDD354 pKa = 3.18FDD356 pKa = 4.22SFSIDD361 pKa = 3.32EE362 pKa = 4.16VSDD365 pKa = 3.66IGSHH369 pKa = 5.96TYY371 pKa = 10.48
MM1 pKa = 7.48KK2 pKa = 10.33KK3 pKa = 9.43ILPFILCNLLALASFAQIGINTDD26 pKa = 3.41SPSPAAALEE35 pKa = 4.33IKK37 pKa = 10.49SSYY40 pKa = 9.4NNKK43 pKa = 10.2NYY45 pKa = 10.69GGLLIPTVDD54 pKa = 2.82ATEE57 pKa = 4.02RR58 pKa = 11.84DD59 pKa = 4.02KK60 pKa = 11.4ISDD63 pKa = 3.58KK64 pKa = 10.92ATTADD69 pKa = 3.53GGMLIYY75 pKa = 10.42YY76 pKa = 9.85VEE78 pKa = 4.53GNTRR82 pKa = 11.84CLQIYY87 pKa = 10.28DD88 pKa = 4.95GINGSWADD96 pKa = 4.03VYY98 pKa = 11.46CMPTNYY104 pKa = 10.42APIASNVFISGIIEE118 pKa = 3.99DD119 pKa = 4.35TQILTANYY127 pKa = 7.87TYY129 pKa = 11.45SDD131 pKa = 3.94IEE133 pKa = 4.06NDD135 pKa = 3.7AEE137 pKa = 4.4GTTTFEE143 pKa = 3.97WFIADD148 pKa = 4.2DD149 pKa = 4.02AQGTNTTSLGVTTTGTYY166 pKa = 9.04TLQTADD172 pKa = 3.02VGKK175 pKa = 10.42FIFVRR180 pKa = 11.84VTPVASAGEE189 pKa = 4.54FIGSPVDD196 pKa = 3.48SPLRR200 pKa = 11.84GPIEE204 pKa = 4.02AAGGVASDD212 pKa = 4.37LFISEE217 pKa = 4.44YY218 pKa = 11.2VEE220 pKa = 3.96GSSYY224 pKa = 11.62NKK226 pKa = 9.94IIEE229 pKa = 4.18IANFTGQDD237 pKa = 3.47VNLSEE242 pKa = 3.87YY243 pKa = 9.94HH244 pKa = 6.49LRR246 pKa = 11.84IFRR249 pKa = 11.84QTGNLTGPISLTSNTLIDD267 pKa = 3.7GDD269 pKa = 4.12VFVIANTANVLATGEE284 pKa = 4.33VDD286 pKa = 3.49TQTGSLNFNGDD297 pKa = 3.78DD298 pKa = 4.07SVLLYY303 pKa = 8.39YY304 pKa = 10.49QPLGGTEE311 pKa = 4.02TLIDD315 pKa = 4.01IIGPDD320 pKa = 3.59SLVNPPSNNPNFAKK334 pKa = 10.72DD335 pKa = 3.12EE336 pKa = 4.17TLRR339 pKa = 11.84RR340 pKa = 11.84KK341 pKa = 9.86PGLGPSTTYY350 pKa = 11.05DD351 pKa = 3.45SNDD354 pKa = 3.18FDD356 pKa = 4.22SFSIDD361 pKa = 3.32EE362 pKa = 4.16VSDD365 pKa = 3.66IGSHH369 pKa = 5.96TYY371 pKa = 10.48
Molecular weight: 39.77 kDa
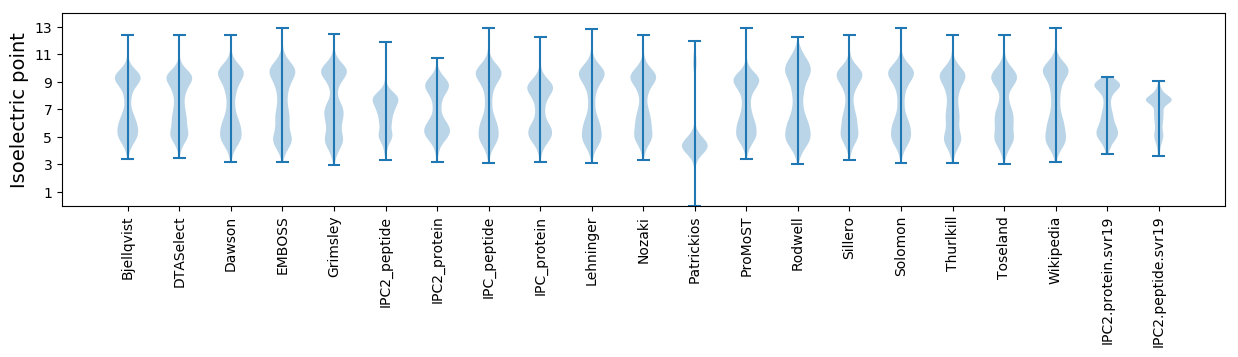
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C8L7H1|A0A5C8L7H1_9FLAO Cytokinin riboside 5'-monophosphate phosphoribohydrolase OS=Mesonia sp. HuA40 OX=2602761 GN=FT993_11195 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.05RR21 pKa = 11.84MASVNGRR28 pKa = 11.84KK29 pKa = 9.21VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.73LSVSSEE47 pKa = 3.76NRR49 pKa = 11.84HH50 pKa = 4.47KK51 pKa = 10.5HH52 pKa = 4.49
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.05RR21 pKa = 11.84MASVNGRR28 pKa = 11.84KK29 pKa = 9.21VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.73LSVSSEE47 pKa = 3.76NRR49 pKa = 11.84HH50 pKa = 4.47KK51 pKa = 10.5HH52 pKa = 4.49
Molecular weight: 6.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
784139 |
32 |
2408 |
329.3 |
37.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.843 ± 0.054 | 0.718 ± 0.017 |
5.212 ± 0.038 | 6.676 ± 0.054 |
5.225 ± 0.04 | 6.048 ± 0.054 |
1.84 ± 0.023 | 7.559 ± 0.048 |
7.962 ± 0.074 | 9.967 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.048 ± 0.024 | 6.078 ± 0.055 |
3.486 ± 0.025 | 4.417 ± 0.037 |
3.554 ± 0.035 | 6.041 ± 0.037 |
5.293 ± 0.05 | 5.931 ± 0.048 |
1.012 ± 0.017 | 4.09 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
