
Torque teno virus (isolate Japanese macaque/Japan/Mf-TTV9/2000) (TTV)
Taxonomy: Viruses; Anelloviridae; Alphatorquevirus
Average proteome isoelectric point is 7.73
Get precalculated fractions of proteins
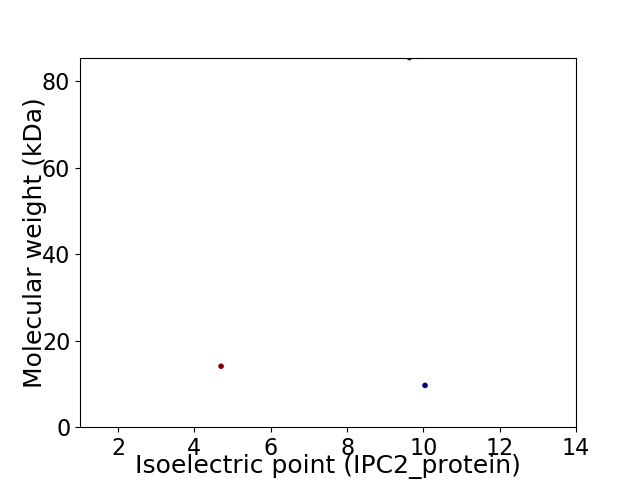
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>sp|Q9DUC5|YORF2_TTVV9 Uncharacterized ORF2 protein OS=Torque teno virus (isolate Japanese macaque/Japan/Mf-TTV9/2000) OX=687364 GN=ORF2 PE=4 SV=1
MM1 pKa = 7.08TNQWEE6 pKa = 4.49PTTLNAWGRR15 pKa = 11.84EE16 pKa = 4.2EE17 pKa = 3.68AWLRR21 pKa = 11.84SVVDD25 pKa = 3.27SHH27 pKa = 7.77QSFCGCNDD35 pKa = 3.53PGFHH39 pKa = 7.06LGLLLHH45 pKa = 6.36SAGRR49 pKa = 11.84HH50 pKa = 4.73LGGPGGPQPPPAPGGPGVGGEE71 pKa = 4.33GPRR74 pKa = 11.84RR75 pKa = 11.84FLPLPGLPANPEE87 pKa = 4.19EE88 pKa = 5.15PGHH91 pKa = 5.79QRR93 pKa = 11.84PPCPGGPGDD102 pKa = 3.93AGGAGGVDD110 pKa = 3.3AEE112 pKa = 4.52TGEE115 pKa = 4.67GEE117 pKa = 4.21WRR119 pKa = 11.84PEE121 pKa = 4.45DD122 pKa = 3.64IAEE125 pKa = 4.48LLDD128 pKa = 3.79GLEE131 pKa = 4.48DD132 pKa = 3.6AEE134 pKa = 4.78KK135 pKa = 10.71RR136 pKa = 3.79
MM1 pKa = 7.08TNQWEE6 pKa = 4.49PTTLNAWGRR15 pKa = 11.84EE16 pKa = 4.2EE17 pKa = 3.68AWLRR21 pKa = 11.84SVVDD25 pKa = 3.27SHH27 pKa = 7.77QSFCGCNDD35 pKa = 3.53PGFHH39 pKa = 7.06LGLLLHH45 pKa = 6.36SAGRR49 pKa = 11.84HH50 pKa = 4.73LGGPGGPQPPPAPGGPGVGGEE71 pKa = 4.33GPRR74 pKa = 11.84RR75 pKa = 11.84FLPLPGLPANPEE87 pKa = 4.19EE88 pKa = 5.15PGHH91 pKa = 5.79QRR93 pKa = 11.84PPCPGGPGDD102 pKa = 3.93AGGAGGVDD110 pKa = 3.3AEE112 pKa = 4.52TGEE115 pKa = 4.67GEE117 pKa = 4.21WRR119 pKa = 11.84PEE121 pKa = 4.45DD122 pKa = 3.64IAEE125 pKa = 4.48LLDD128 pKa = 3.79GLEE131 pKa = 4.48DD132 pKa = 3.6AEE134 pKa = 4.78KK135 pKa = 10.71RR136 pKa = 3.79
Molecular weight: 14.09 kDa
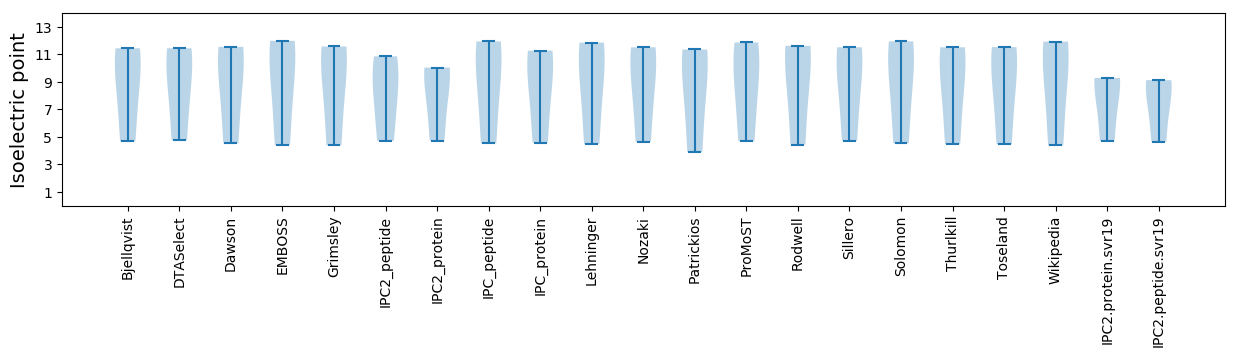
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9DUC5|YORF2_TTVV9 Uncharacterized ORF2 protein OS=Torque teno virus (isolate Japanese macaque/Japan/Mf-TTV9/2000) OX=687364 GN=ORF2 PE=4 SV=1
MM1 pKa = 7.56PWWPWRR7 pKa = 11.84RR8 pKa = 11.84WRR10 pKa = 11.84RR11 pKa = 11.84WRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84GNWRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84VAPRR26 pKa = 11.84RR27 pKa = 11.84YY28 pKa = 9.42RR29 pKa = 11.84RR30 pKa = 11.84TARR33 pKa = 11.84RR34 pKa = 11.84ARR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 8.55AVRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84VRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84GYY55 pKa = 7.43YY56 pKa = 8.41RR57 pKa = 11.84RR58 pKa = 11.84YY59 pKa = 9.0RR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84YY65 pKa = 9.64KK66 pKa = 10.18RR67 pKa = 11.84RR68 pKa = 11.84LLILKK73 pKa = 8.65QWQPTTNLRR82 pKa = 11.84LTVKK86 pKa = 10.46GLIPVVVMGKK96 pKa = 10.63GKK98 pKa = 7.51TQNNFGQWEE107 pKa = 4.14QTVPLEE113 pKa = 4.26GEE115 pKa = 4.33SYY117 pKa = 10.84GGSFTIRR124 pKa = 11.84KK125 pKa = 7.25FTLQTLYY132 pKa = 11.08EE133 pKa = 5.06DD134 pKa = 3.85YY135 pKa = 11.35LKK137 pKa = 10.97LRR139 pKa = 11.84NRR141 pKa = 11.84WSRR144 pKa = 11.84SNTDD148 pKa = 3.38LEE150 pKa = 4.73LIRR153 pKa = 11.84YY154 pKa = 6.71TGLNLRR160 pKa = 11.84LYY162 pKa = 9.52RR163 pKa = 11.84HH164 pKa = 6.19EE165 pKa = 4.22FSDD168 pKa = 4.59YY169 pKa = 9.26IVHH172 pKa = 6.45YY173 pKa = 10.48SLEE176 pKa = 4.29TPMEE180 pKa = 4.39VGLEE184 pKa = 4.1SHH186 pKa = 6.51MLAHH190 pKa = 6.84PLKK193 pKa = 9.87MIMSSKK199 pKa = 10.28HH200 pKa = 3.47VTVPSLLTKK209 pKa = 10.41KK210 pKa = 10.45GGRR213 pKa = 11.84RR214 pKa = 11.84YY215 pKa = 10.07LRR217 pKa = 11.84LKK219 pKa = 10.38IPPPKK224 pKa = 10.73LMMTQWYY231 pKa = 7.62FQKK234 pKa = 10.44EE235 pKa = 4.18FCQVGLVLLSISTATLMHH253 pKa = 6.97PWMAPFVNSPALTIYY268 pKa = 10.53AVNHH272 pKa = 5.13KK273 pKa = 8.76TYY275 pKa = 10.95SDD277 pKa = 3.6MSILPSNNQGSTKK290 pKa = 10.67NEE292 pKa = 4.45LIEE295 pKa = 4.04TLYY298 pKa = 10.24TAEE301 pKa = 3.93HH302 pKa = 6.85TYY304 pKa = 11.39NPMAQRR310 pKa = 11.84IWGNIKK316 pKa = 9.45PQNTNFTPEE325 pKa = 4.11NYY327 pKa = 7.74WNKK330 pKa = 9.18WNEE333 pKa = 3.33IHH335 pKa = 7.15TKK337 pKa = 10.09IKK339 pKa = 9.91TNRR342 pKa = 11.84QSEE345 pKa = 4.4LTQLKK350 pKa = 9.35QMRR353 pKa = 11.84KK354 pKa = 8.82RR355 pKa = 11.84LEE357 pKa = 3.93LTTEE361 pKa = 3.98NDD363 pKa = 3.48YY364 pKa = 11.36QDD366 pKa = 4.04PNFGLSYY373 pKa = 11.09GLYY376 pKa = 10.2SPLLLYY382 pKa = 10.43PEE384 pKa = 4.91MYY386 pKa = 10.32FPEE389 pKa = 4.1QSKK392 pKa = 10.38VYY394 pKa = 10.48QKK396 pKa = 11.26ARR398 pKa = 11.84YY399 pKa = 8.99NPLLDD404 pKa = 3.43QGIGNVVWTEE414 pKa = 3.67PLTKK418 pKa = 9.38KK419 pKa = 8.16TCDD422 pKa = 3.72YY423 pKa = 11.34ASQAYY428 pKa = 9.38NVIKK432 pKa = 10.44DD433 pKa = 3.63APLYY437 pKa = 10.25LALFGYY443 pKa = 10.22IDD445 pKa = 3.86ICSKK449 pKa = 10.19LAKK452 pKa = 10.18DD453 pKa = 3.32KK454 pKa = 11.38SFYY457 pKa = 10.58LSNRR461 pKa = 11.84VCVKK465 pKa = 10.4CPYY468 pKa = 9.18TVPQLLSKK476 pKa = 9.82TNATLGHH483 pKa = 6.38VILSEE488 pKa = 3.75NFMRR492 pKa = 11.84GLVPSKK498 pKa = 10.71DD499 pKa = 3.79SYY501 pKa = 10.85VPLYY505 pKa = 10.22MRR507 pKa = 11.84GKK509 pKa = 8.43WYY511 pKa = 9.56PSIYY515 pKa = 9.48HH516 pKa = 4.83QEE518 pKa = 3.98EE519 pKa = 4.21VIEE522 pKa = 5.21AIVSSGPFVPRR533 pKa = 11.84DD534 pKa = 3.7QITKK538 pKa = 10.48SLDD541 pKa = 2.95ITIGCRR547 pKa = 11.84FGFRR551 pKa = 11.84IGGNLLNPKK560 pKa = 9.77QVGDD564 pKa = 3.63PCKK567 pKa = 10.47QPTHH571 pKa = 6.96PLPAPGGGDD580 pKa = 3.71LLRR583 pKa = 11.84AVQVSDD589 pKa = 3.41PRR591 pKa = 11.84KK592 pKa = 10.09VGIQFHH598 pKa = 6.34PWDD601 pKa = 4.34LRR603 pKa = 11.84RR604 pKa = 11.84GMLSTSSIKK613 pKa = 10.58RR614 pKa = 11.84MLQDD618 pKa = 3.36SDD620 pKa = 4.27DD621 pKa = 4.52DD622 pKa = 4.29EE623 pKa = 6.82SIEE626 pKa = 4.38FPPKK630 pKa = 10.08AWPGDD635 pKa = 3.66PVPVGRR641 pKa = 11.84TLEE644 pKa = 4.19EE645 pKa = 4.08RR646 pKa = 11.84CSSSLYY652 pKa = 10.64HH653 pKa = 7.08LLQEE657 pKa = 4.58QATPPPFKK665 pKa = 10.59KK666 pKa = 10.05PRR668 pKa = 11.84TEE670 pKa = 3.94DD671 pKa = 3.19QEE673 pKa = 4.38EE674 pKa = 4.27NPEE677 pKa = 4.07EE678 pKa = 4.2TTQLQLFQEE687 pKa = 4.63LQRR690 pKa = 11.84QRR692 pKa = 11.84EE693 pKa = 4.11LQLQLKK699 pKa = 9.86RR700 pKa = 11.84GFRR703 pKa = 11.84GLVEE707 pKa = 4.32EE708 pKa = 4.65MIKK711 pKa = 9.06SHH713 pKa = 6.32RR714 pKa = 11.84HH715 pKa = 4.74LALDD719 pKa = 4.69PYY721 pKa = 11.19LKK723 pKa = 10.77
MM1 pKa = 7.56PWWPWRR7 pKa = 11.84RR8 pKa = 11.84WRR10 pKa = 11.84RR11 pKa = 11.84WRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84GNWRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84VAPRR26 pKa = 11.84RR27 pKa = 11.84YY28 pKa = 9.42RR29 pKa = 11.84RR30 pKa = 11.84TARR33 pKa = 11.84RR34 pKa = 11.84ARR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 8.55AVRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84VRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84GYY55 pKa = 7.43YY56 pKa = 8.41RR57 pKa = 11.84RR58 pKa = 11.84YY59 pKa = 9.0RR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84YY65 pKa = 9.64KK66 pKa = 10.18RR67 pKa = 11.84RR68 pKa = 11.84LLILKK73 pKa = 8.65QWQPTTNLRR82 pKa = 11.84LTVKK86 pKa = 10.46GLIPVVVMGKK96 pKa = 10.63GKK98 pKa = 7.51TQNNFGQWEE107 pKa = 4.14QTVPLEE113 pKa = 4.26GEE115 pKa = 4.33SYY117 pKa = 10.84GGSFTIRR124 pKa = 11.84KK125 pKa = 7.25FTLQTLYY132 pKa = 11.08EE133 pKa = 5.06DD134 pKa = 3.85YY135 pKa = 11.35LKK137 pKa = 10.97LRR139 pKa = 11.84NRR141 pKa = 11.84WSRR144 pKa = 11.84SNTDD148 pKa = 3.38LEE150 pKa = 4.73LIRR153 pKa = 11.84YY154 pKa = 6.71TGLNLRR160 pKa = 11.84LYY162 pKa = 9.52RR163 pKa = 11.84HH164 pKa = 6.19EE165 pKa = 4.22FSDD168 pKa = 4.59YY169 pKa = 9.26IVHH172 pKa = 6.45YY173 pKa = 10.48SLEE176 pKa = 4.29TPMEE180 pKa = 4.39VGLEE184 pKa = 4.1SHH186 pKa = 6.51MLAHH190 pKa = 6.84PLKK193 pKa = 9.87MIMSSKK199 pKa = 10.28HH200 pKa = 3.47VTVPSLLTKK209 pKa = 10.41KK210 pKa = 10.45GGRR213 pKa = 11.84RR214 pKa = 11.84YY215 pKa = 10.07LRR217 pKa = 11.84LKK219 pKa = 10.38IPPPKK224 pKa = 10.73LMMTQWYY231 pKa = 7.62FQKK234 pKa = 10.44EE235 pKa = 4.18FCQVGLVLLSISTATLMHH253 pKa = 6.97PWMAPFVNSPALTIYY268 pKa = 10.53AVNHH272 pKa = 5.13KK273 pKa = 8.76TYY275 pKa = 10.95SDD277 pKa = 3.6MSILPSNNQGSTKK290 pKa = 10.67NEE292 pKa = 4.45LIEE295 pKa = 4.04TLYY298 pKa = 10.24TAEE301 pKa = 3.93HH302 pKa = 6.85TYY304 pKa = 11.39NPMAQRR310 pKa = 11.84IWGNIKK316 pKa = 9.45PQNTNFTPEE325 pKa = 4.11NYY327 pKa = 7.74WNKK330 pKa = 9.18WNEE333 pKa = 3.33IHH335 pKa = 7.15TKK337 pKa = 10.09IKK339 pKa = 9.91TNRR342 pKa = 11.84QSEE345 pKa = 4.4LTQLKK350 pKa = 9.35QMRR353 pKa = 11.84KK354 pKa = 8.82RR355 pKa = 11.84LEE357 pKa = 3.93LTTEE361 pKa = 3.98NDD363 pKa = 3.48YY364 pKa = 11.36QDD366 pKa = 4.04PNFGLSYY373 pKa = 11.09GLYY376 pKa = 10.2SPLLLYY382 pKa = 10.43PEE384 pKa = 4.91MYY386 pKa = 10.32FPEE389 pKa = 4.1QSKK392 pKa = 10.38VYY394 pKa = 10.48QKK396 pKa = 11.26ARR398 pKa = 11.84YY399 pKa = 8.99NPLLDD404 pKa = 3.43QGIGNVVWTEE414 pKa = 3.67PLTKK418 pKa = 9.38KK419 pKa = 8.16TCDD422 pKa = 3.72YY423 pKa = 11.34ASQAYY428 pKa = 9.38NVIKK432 pKa = 10.44DD433 pKa = 3.63APLYY437 pKa = 10.25LALFGYY443 pKa = 10.22IDD445 pKa = 3.86ICSKK449 pKa = 10.19LAKK452 pKa = 10.18DD453 pKa = 3.32KK454 pKa = 11.38SFYY457 pKa = 10.58LSNRR461 pKa = 11.84VCVKK465 pKa = 10.4CPYY468 pKa = 9.18TVPQLLSKK476 pKa = 9.82TNATLGHH483 pKa = 6.38VILSEE488 pKa = 3.75NFMRR492 pKa = 11.84GLVPSKK498 pKa = 10.71DD499 pKa = 3.79SYY501 pKa = 10.85VPLYY505 pKa = 10.22MRR507 pKa = 11.84GKK509 pKa = 8.43WYY511 pKa = 9.56PSIYY515 pKa = 9.48HH516 pKa = 4.83QEE518 pKa = 3.98EE519 pKa = 4.21VIEE522 pKa = 5.21AIVSSGPFVPRR533 pKa = 11.84DD534 pKa = 3.7QITKK538 pKa = 10.48SLDD541 pKa = 2.95ITIGCRR547 pKa = 11.84FGFRR551 pKa = 11.84IGGNLLNPKK560 pKa = 9.77QVGDD564 pKa = 3.63PCKK567 pKa = 10.47QPTHH571 pKa = 6.96PLPAPGGGDD580 pKa = 3.71LLRR583 pKa = 11.84AVQVSDD589 pKa = 3.41PRR591 pKa = 11.84KK592 pKa = 10.09VGIQFHH598 pKa = 6.34PWDD601 pKa = 4.34LRR603 pKa = 11.84RR604 pKa = 11.84GMLSTSSIKK613 pKa = 10.58RR614 pKa = 11.84MLQDD618 pKa = 3.36SDD620 pKa = 4.27DD621 pKa = 4.52DD622 pKa = 4.29EE623 pKa = 6.82SIEE626 pKa = 4.38FPPKK630 pKa = 10.08AWPGDD635 pKa = 3.66PVPVGRR641 pKa = 11.84TLEE644 pKa = 4.19EE645 pKa = 4.08RR646 pKa = 11.84CSSSLYY652 pKa = 10.64HH653 pKa = 7.08LLQEE657 pKa = 4.58QATPPPFKK665 pKa = 10.59KK666 pKa = 10.05PRR668 pKa = 11.84TEE670 pKa = 3.94DD671 pKa = 3.19QEE673 pKa = 4.38EE674 pKa = 4.27NPEE677 pKa = 4.07EE678 pKa = 4.2TTQLQLFQEE687 pKa = 4.63LQRR690 pKa = 11.84QRR692 pKa = 11.84EE693 pKa = 4.11LQLQLKK699 pKa = 9.86RR700 pKa = 11.84GFRR703 pKa = 11.84GLVEE707 pKa = 4.32EE708 pKa = 4.65MIKK711 pKa = 9.06SHH713 pKa = 6.32RR714 pKa = 11.84HH715 pKa = 4.74LALDD719 pKa = 4.69PYY721 pKa = 11.19LKK723 pKa = 10.77
Molecular weight: 85.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
945 |
86 |
723 |
315.0 |
36.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.81 ± 1.197 | 1.27 ± 0.326 |
3.598 ± 0.547 | 5.926 ± 1.23 |
2.54 ± 0.719 | 7.407 ± 4.262 |
2.328 ± 0.481 | 3.492 ± 1.318 |
6.032 ± 1.99 | 10.37 ± 1.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.434 ± 0.58 | 4.021 ± 0.451 |
7.937 ± 2.321 | 4.868 ± 0.708 |
9.947 ± 1.402 | 7.407 ± 4.841 |
5.608 ± 1.061 | 4.444 ± 0.892 |
2.434 ± 0.326 | 4.127 ± 1.878 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
