
Phasianus colchicus (Common pheasant)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Sauropsida; Sauria; Archelosauria; Archosauria; Dinosauria;
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
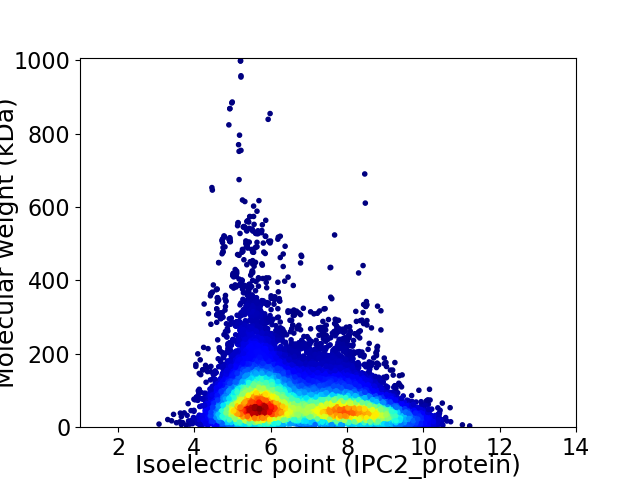
Virtual 2D-PAGE plot for 25507 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A669Q1U1|A0A669Q1U1_PHACC SPARC (osteonectin) cwcv and kazal like domains proteoglycan 1 OS=Phasianus colchicus OX=9054 PE=4 SV=1
MM1 pKa = 7.43SSSLGRR7 pKa = 11.84STTWVRR13 pKa = 11.84PSRR16 pKa = 11.84APCPGPAMPPGTQSSARR33 pKa = 11.84SAALSRR39 pKa = 11.84DD40 pKa = 3.53GAGDD44 pKa = 3.49PGMWIPQGQALPAAGEE60 pKa = 4.29GAAEE64 pKa = 3.96QAGAGARR71 pKa = 11.84EE72 pKa = 4.47GWTGVMMMVVMVGVMVGMVMVGGDD96 pKa = 4.14DD97 pKa = 5.0DD98 pKa = 6.47DD99 pKa = 7.27DD100 pKa = 6.09GFDD103 pKa = 4.05SDD105 pKa = 5.77GGGDD109 pKa = 4.32DD110 pKa = 5.32DD111 pKa = 5.71DD112 pKa = 6.31GSGGSVVDD120 pKa = 4.82GDD122 pKa = 5.66DD123 pKa = 3.56VDD125 pKa = 6.0DD126 pKa = 6.08DD127 pKa = 5.11GGDD130 pKa = 3.54VDD132 pKa = 6.37DD133 pKa = 6.36GDD135 pKa = 6.01DD136 pKa = 5.0GDD138 pKa = 5.52DD139 pKa = 3.86VDD141 pKa = 6.6DD142 pKa = 6.71GGDD145 pKa = 3.69DD146 pKa = 4.46DD147 pKa = 6.07NDD149 pKa = 4.11GGGDD153 pKa = 3.91DD154 pKa = 5.86DD155 pKa = 6.54GGGDD159 pKa = 5.68DD160 pKa = 6.6DD161 pKa = 4.91GTLQNASGSATDD173 pKa = 4.14ASASEE178 pKa = 4.48AFWNEE183 pKa = 3.31QVLGVTHH190 pKa = 6.98SSGLGDD196 pKa = 3.93PGSVQWLLALCLLAAWLVVFLCMLRR221 pKa = 11.84GIHH224 pKa = 6.69SSGKK228 pKa = 9.38VVYY231 pKa = 8.85ITATFPYY238 pKa = 10.21LILLILIIRR247 pKa = 11.84GATLPGSLDD256 pKa = 3.18GVRR259 pKa = 11.84FYY261 pKa = 11.48LSSDD265 pKa = 2.67WSKK268 pKa = 11.28LLSAQVWSDD277 pKa = 2.9AASQIFYY284 pKa = 10.98SLGIGFGGLLSMASYY299 pKa = 11.56NKK301 pKa = 9.66FDD303 pKa = 4.22NNVVRR308 pKa = 11.84DD309 pKa = 3.88TLVIAVGNCCTSFFAGFAIFSVLGHH334 pKa = 6.21MALKK338 pKa = 10.34RR339 pKa = 11.84DD340 pKa = 3.54IPVGSVAEE348 pKa = 4.35SGPGLAFVAYY358 pKa = 9.43PEE360 pKa = 4.58ALSLLPGSPFWSILFFLMLFMLGVDD385 pKa = 3.52TLFGNIEE392 pKa = 4.85AITTAIMDD400 pKa = 4.18EE401 pKa = 4.63FPALRR406 pKa = 11.84EE407 pKa = 4.02GRR409 pKa = 11.84RR410 pKa = 11.84KK411 pKa = 10.47VMLLAVLCFSFFLLGLLLVTQGGIFWFTLIDD442 pKa = 3.63TYY444 pKa = 11.09STGFGLIIITLLMCIGIASCYY465 pKa = 10.0GIEE468 pKa = 4.24RR469 pKa = 11.84FCRR472 pKa = 11.84DD473 pKa = 3.74IVTMICLCPPWYY485 pKa = 10.42SRR487 pKa = 11.84VLGCFKK493 pKa = 10.68VCWVFFTPCLLLFTLIFTFLDD514 pKa = 3.77MYY516 pKa = 10.68NIPLSYY522 pKa = 9.43GTYY525 pKa = 10.12EE526 pKa = 4.1FPAWGTSLGVCMGVLSCVQIPIGAIVALCHH556 pKa = 5.06QTGTLTDD563 pKa = 4.07RR564 pKa = 11.84FQKK567 pKa = 10.68AMQPLHH573 pKa = 6.0SWGTAAASAEE583 pKa = 4.48DD584 pKa = 3.94IIEE587 pKa = 4.23VPFTITLSSSDD598 pKa = 3.82FTTPPHH604 pKa = 6.6SEE606 pKa = 3.97SS607 pKa = 3.4
MM1 pKa = 7.43SSSLGRR7 pKa = 11.84STTWVRR13 pKa = 11.84PSRR16 pKa = 11.84APCPGPAMPPGTQSSARR33 pKa = 11.84SAALSRR39 pKa = 11.84DD40 pKa = 3.53GAGDD44 pKa = 3.49PGMWIPQGQALPAAGEE60 pKa = 4.29GAAEE64 pKa = 3.96QAGAGARR71 pKa = 11.84EE72 pKa = 4.47GWTGVMMMVVMVGVMVGMVMVGGDD96 pKa = 4.14DD97 pKa = 5.0DD98 pKa = 6.47DD99 pKa = 7.27DD100 pKa = 6.09GFDD103 pKa = 4.05SDD105 pKa = 5.77GGGDD109 pKa = 4.32DD110 pKa = 5.32DD111 pKa = 5.71DD112 pKa = 6.31GSGGSVVDD120 pKa = 4.82GDD122 pKa = 5.66DD123 pKa = 3.56VDD125 pKa = 6.0DD126 pKa = 6.08DD127 pKa = 5.11GGDD130 pKa = 3.54VDD132 pKa = 6.37DD133 pKa = 6.36GDD135 pKa = 6.01DD136 pKa = 5.0GDD138 pKa = 5.52DD139 pKa = 3.86VDD141 pKa = 6.6DD142 pKa = 6.71GGDD145 pKa = 3.69DD146 pKa = 4.46DD147 pKa = 6.07NDD149 pKa = 4.11GGGDD153 pKa = 3.91DD154 pKa = 5.86DD155 pKa = 6.54GGGDD159 pKa = 5.68DD160 pKa = 6.6DD161 pKa = 4.91GTLQNASGSATDD173 pKa = 4.14ASASEE178 pKa = 4.48AFWNEE183 pKa = 3.31QVLGVTHH190 pKa = 6.98SSGLGDD196 pKa = 3.93PGSVQWLLALCLLAAWLVVFLCMLRR221 pKa = 11.84GIHH224 pKa = 6.69SSGKK228 pKa = 9.38VVYY231 pKa = 8.85ITATFPYY238 pKa = 10.21LILLILIIRR247 pKa = 11.84GATLPGSLDD256 pKa = 3.18GVRR259 pKa = 11.84FYY261 pKa = 11.48LSSDD265 pKa = 2.67WSKK268 pKa = 11.28LLSAQVWSDD277 pKa = 2.9AASQIFYY284 pKa = 10.98SLGIGFGGLLSMASYY299 pKa = 11.56NKK301 pKa = 9.66FDD303 pKa = 4.22NNVVRR308 pKa = 11.84DD309 pKa = 3.88TLVIAVGNCCTSFFAGFAIFSVLGHH334 pKa = 6.21MALKK338 pKa = 10.34RR339 pKa = 11.84DD340 pKa = 3.54IPVGSVAEE348 pKa = 4.35SGPGLAFVAYY358 pKa = 9.43PEE360 pKa = 4.58ALSLLPGSPFWSILFFLMLFMLGVDD385 pKa = 3.52TLFGNIEE392 pKa = 4.85AITTAIMDD400 pKa = 4.18EE401 pKa = 4.63FPALRR406 pKa = 11.84EE407 pKa = 4.02GRR409 pKa = 11.84RR410 pKa = 11.84KK411 pKa = 10.47VMLLAVLCFSFFLLGLLLVTQGGIFWFTLIDD442 pKa = 3.63TYY444 pKa = 11.09STGFGLIIITLLMCIGIASCYY465 pKa = 10.0GIEE468 pKa = 4.24RR469 pKa = 11.84FCRR472 pKa = 11.84DD473 pKa = 3.74IVTMICLCPPWYY485 pKa = 10.42SRR487 pKa = 11.84VLGCFKK493 pKa = 10.68VCWVFFTPCLLLFTLIFTFLDD514 pKa = 3.77MYY516 pKa = 10.68NIPLSYY522 pKa = 9.43GTYY525 pKa = 10.12EE526 pKa = 4.1FPAWGTSLGVCMGVLSCVQIPIGAIVALCHH556 pKa = 5.06QTGTLTDD563 pKa = 4.07RR564 pKa = 11.84FQKK567 pKa = 10.68AMQPLHH573 pKa = 6.0SWGTAAASAEE583 pKa = 4.48DD584 pKa = 3.94IIEE587 pKa = 4.23VPFTITLSSSDD598 pKa = 3.82FTTPPHH604 pKa = 6.6SEE606 pKa = 3.97SS607 pKa = 3.4
Molecular weight: 64.15 kDa
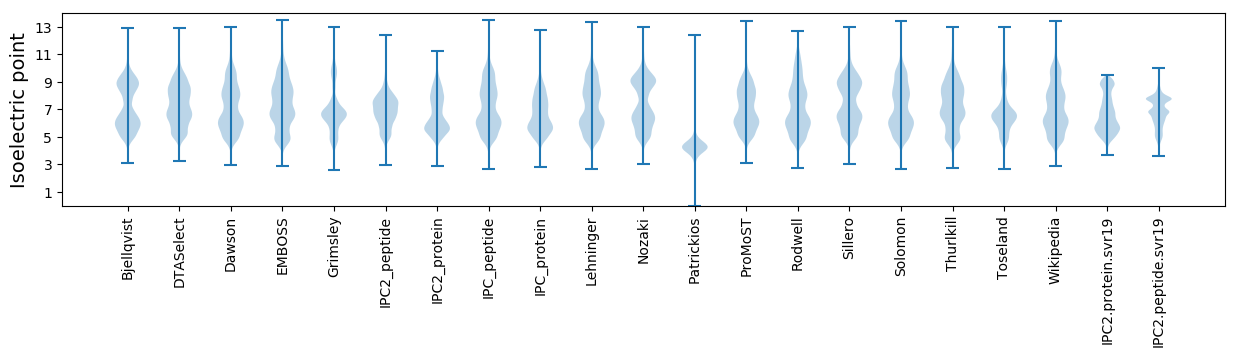
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A669QY88|A0A669QY88_PHACC Cyclin L2 OS=Phasianus colchicus OX=9054 PE=3 SV=1
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.12WRR6 pKa = 11.84KK7 pKa = 9.1KK8 pKa = 9.32RR9 pKa = 11.84MRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 10.08RR15 pKa = 11.84KK16 pKa = 8.21RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.46MRR21 pKa = 11.84QRR23 pKa = 11.84SKK25 pKa = 11.41
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.12WRR6 pKa = 11.84KK7 pKa = 9.1KK8 pKa = 9.32RR9 pKa = 11.84MRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 10.08RR15 pKa = 11.84KK16 pKa = 8.21RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.46MRR21 pKa = 11.84QRR23 pKa = 11.84SKK25 pKa = 11.41
Molecular weight: 3.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
17908591 |
25 |
8758 |
702.1 |
78.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.837 ± 0.015 | 2.302 ± 0.015 |
4.974 ± 0.01 | 7.202 ± 0.019 |
3.7 ± 0.01 | 6.194 ± 0.018 |
2.529 ± 0.007 | 4.758 ± 0.011 |
6.052 ± 0.017 | 9.684 ± 0.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.243 ± 0.006 | 3.972 ± 0.01 |
5.608 ± 0.019 | 4.721 ± 0.014 |
5.448 ± 0.014 | 8.324 ± 0.017 |
5.308 ± 0.01 | 6.121 ± 0.01 |
1.201 ± 0.005 | 2.792 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
