
Tenacibaculum phage JQ
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
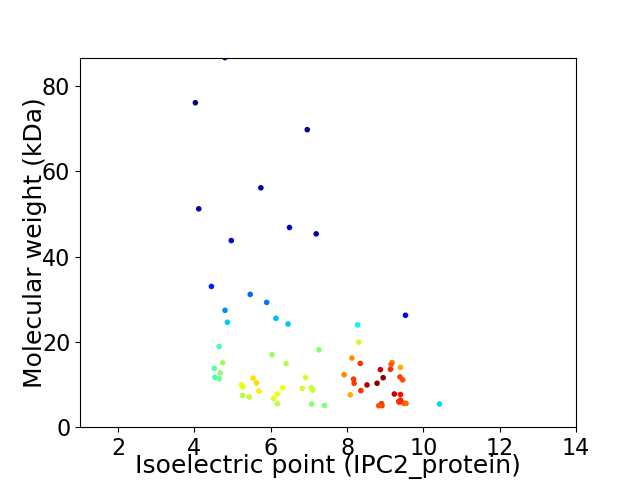
Virtual 2D-PAGE plot for 72 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G6XUV2|A0A6G6XUV2_9CAUD Uncharacterized protein OS=Tenacibaculum phage JQ OX=2712935 PE=4 SV=1
MM1 pKa = 7.79ASNIIIQFNSALQSGDD17 pKa = 3.47FLRR20 pKa = 11.84LQDD23 pKa = 3.74SYY25 pKa = 11.37TPNVEE30 pKa = 3.83MGFYY34 pKa = 9.29WGSASGFLPVTGNISSDD51 pKa = 2.83INNLKK56 pKa = 10.31NRR58 pKa = 11.84INTAYY63 pKa = 10.65NSTNRR68 pKa = 11.84YY69 pKa = 6.18TVTSDD74 pKa = 2.79ATSITITDD82 pKa = 3.74NIGFSEE88 pKa = 4.51FSEE91 pKa = 4.66VNNSTGARR99 pKa = 11.84LDD101 pKa = 3.27ITIVNDD107 pKa = 3.44SQQIGVNIDD116 pKa = 3.55NIEE119 pKa = 4.43LIEE122 pKa = 4.34NATNPCDD129 pKa = 3.68LVDD132 pKa = 4.51VRR134 pKa = 11.84ITTNEE139 pKa = 3.68APFDD143 pKa = 3.94LLQPVSVTDD152 pKa = 3.59IQTTTYY158 pKa = 9.81TISDD162 pKa = 3.77VQRR165 pKa = 11.84SINFINVEE173 pKa = 4.18VKK175 pKa = 10.4KK176 pKa = 11.29GNITDD181 pKa = 3.97LEE183 pKa = 4.46EE184 pKa = 4.39IFAPVINSEE193 pKa = 4.17LFKK196 pKa = 10.92LDD198 pKa = 3.41VVNTPSGATLSIRR211 pKa = 11.84WKK213 pKa = 10.19GVRR216 pKa = 11.84NPFFDD221 pKa = 4.63LEE223 pKa = 4.1YY224 pKa = 10.51SINGVDD230 pKa = 4.16YY231 pKa = 11.03YY232 pKa = 11.51SSTSFSGLEE241 pKa = 3.95DD242 pKa = 3.45GNYY245 pKa = 9.02TLYY248 pKa = 10.4IRR250 pKa = 11.84DD251 pKa = 4.27SIGCSITIPFEE262 pKa = 4.0VTEE265 pKa = 4.39FEE267 pKa = 4.67PNVYY271 pKa = 10.11EE272 pKa = 3.77RR273 pKa = 11.84TSFFEE278 pKa = 4.04ISEE281 pKa = 4.2QNSNIWIKK289 pKa = 10.15QEE291 pKa = 4.05YY292 pKa = 9.71KK293 pKa = 10.67DD294 pKa = 3.66SCNVYY299 pKa = 10.9SNLTNTLSYY308 pKa = 11.58EE309 pKa = 3.89EE310 pKa = 4.2TTQVNNQSFKK320 pKa = 11.19QLFQKK325 pKa = 10.77CDD327 pKa = 3.91GIVTNQYY334 pKa = 7.53KK335 pKa = 9.73TNYY338 pKa = 8.73EE339 pKa = 3.87NVEE342 pKa = 4.08IKK344 pKa = 10.89LIDD347 pKa = 3.81CDD349 pKa = 4.21GNEE352 pKa = 4.22SLLVSNQISQNINITDD368 pKa = 3.47VRR370 pKa = 11.84DD371 pKa = 3.78VTIEE375 pKa = 3.78AYY377 pKa = 9.64EE378 pKa = 4.01YY379 pKa = 10.94NEE381 pKa = 4.1SSFVGVTYY389 pKa = 10.93VSGNTYY395 pKa = 11.03DD396 pKa = 4.72SNTLTANGSYY406 pKa = 10.57YY407 pKa = 10.53LASSVPSFMNIDD419 pKa = 3.71DD420 pKa = 5.53FIQIEE425 pKa = 4.29GAGWYY430 pKa = 9.21RR431 pKa = 11.84VRR433 pKa = 11.84DD434 pKa = 3.5IIFYY438 pKa = 11.28NNIQTLILDD447 pKa = 3.82VLEE450 pKa = 4.56SDD452 pKa = 5.25FPISLNQTVKK462 pKa = 9.73GTSIYY467 pKa = 10.45NQLDD471 pKa = 3.66YY472 pKa = 11.28EE473 pKa = 4.51LYY475 pKa = 9.39EE476 pKa = 4.06ASYY479 pKa = 11.39DD480 pKa = 3.94LSTLNGDD487 pKa = 3.75YY488 pKa = 11.0YY489 pKa = 10.92LTLDD493 pKa = 3.74ATDD496 pKa = 4.2SEE498 pKa = 5.07FDD500 pKa = 3.61TVSYY504 pKa = 10.19RR505 pKa = 11.84SEE507 pKa = 3.94WFNVASMQYY516 pKa = 8.5NTYY519 pKa = 10.72LLQYY523 pKa = 9.93YY524 pKa = 10.19NSEE527 pKa = 4.25NNEE530 pKa = 4.01TNYY533 pKa = 11.03SNGIRR538 pKa = 11.84NKK540 pKa = 10.1FRR542 pKa = 11.84IRR544 pKa = 11.84YY545 pKa = 7.38QKK547 pKa = 10.33QFTYY551 pKa = 10.4IPNDD555 pKa = 3.47TNEE558 pKa = 4.12VYY560 pKa = 9.5LTDD563 pKa = 3.75TNAVQTDD570 pKa = 3.55SQYY573 pKa = 11.38RR574 pKa = 11.84DD575 pKa = 3.69YY576 pKa = 11.66LKK578 pKa = 10.74LDD580 pKa = 4.15LMPMPVNMVRR590 pKa = 11.84KK591 pKa = 10.15LGLAFSNDD599 pKa = 2.74RR600 pKa = 11.84VFINGLSLIKK610 pKa = 10.82NNEE613 pKa = 4.05LEE615 pKa = 4.06VEE617 pKa = 4.53RR618 pKa = 11.84IGEE621 pKa = 4.16SNEE624 pKa = 3.66YY625 pKa = 10.03RR626 pKa = 11.84FSAQFVRR633 pKa = 11.84SDD635 pKa = 3.31YY636 pKa = 11.53AFDD639 pKa = 3.54VTISDD644 pKa = 3.7NSIVLPQGQPLQINDD659 pKa = 3.52NAEE662 pKa = 3.74GLLFINN668 pKa = 5.04
MM1 pKa = 7.79ASNIIIQFNSALQSGDD17 pKa = 3.47FLRR20 pKa = 11.84LQDD23 pKa = 3.74SYY25 pKa = 11.37TPNVEE30 pKa = 3.83MGFYY34 pKa = 9.29WGSASGFLPVTGNISSDD51 pKa = 2.83INNLKK56 pKa = 10.31NRR58 pKa = 11.84INTAYY63 pKa = 10.65NSTNRR68 pKa = 11.84YY69 pKa = 6.18TVTSDD74 pKa = 2.79ATSITITDD82 pKa = 3.74NIGFSEE88 pKa = 4.51FSEE91 pKa = 4.66VNNSTGARR99 pKa = 11.84LDD101 pKa = 3.27ITIVNDD107 pKa = 3.44SQQIGVNIDD116 pKa = 3.55NIEE119 pKa = 4.43LIEE122 pKa = 4.34NATNPCDD129 pKa = 3.68LVDD132 pKa = 4.51VRR134 pKa = 11.84ITTNEE139 pKa = 3.68APFDD143 pKa = 3.94LLQPVSVTDD152 pKa = 3.59IQTTTYY158 pKa = 9.81TISDD162 pKa = 3.77VQRR165 pKa = 11.84SINFINVEE173 pKa = 4.18VKK175 pKa = 10.4KK176 pKa = 11.29GNITDD181 pKa = 3.97LEE183 pKa = 4.46EE184 pKa = 4.39IFAPVINSEE193 pKa = 4.17LFKK196 pKa = 10.92LDD198 pKa = 3.41VVNTPSGATLSIRR211 pKa = 11.84WKK213 pKa = 10.19GVRR216 pKa = 11.84NPFFDD221 pKa = 4.63LEE223 pKa = 4.1YY224 pKa = 10.51SINGVDD230 pKa = 4.16YY231 pKa = 11.03YY232 pKa = 11.51SSTSFSGLEE241 pKa = 3.95DD242 pKa = 3.45GNYY245 pKa = 9.02TLYY248 pKa = 10.4IRR250 pKa = 11.84DD251 pKa = 4.27SIGCSITIPFEE262 pKa = 4.0VTEE265 pKa = 4.39FEE267 pKa = 4.67PNVYY271 pKa = 10.11EE272 pKa = 3.77RR273 pKa = 11.84TSFFEE278 pKa = 4.04ISEE281 pKa = 4.2QNSNIWIKK289 pKa = 10.15QEE291 pKa = 4.05YY292 pKa = 9.71KK293 pKa = 10.67DD294 pKa = 3.66SCNVYY299 pKa = 10.9SNLTNTLSYY308 pKa = 11.58EE309 pKa = 3.89EE310 pKa = 4.2TTQVNNQSFKK320 pKa = 11.19QLFQKK325 pKa = 10.77CDD327 pKa = 3.91GIVTNQYY334 pKa = 7.53KK335 pKa = 9.73TNYY338 pKa = 8.73EE339 pKa = 3.87NVEE342 pKa = 4.08IKK344 pKa = 10.89LIDD347 pKa = 3.81CDD349 pKa = 4.21GNEE352 pKa = 4.22SLLVSNQISQNINITDD368 pKa = 3.47VRR370 pKa = 11.84DD371 pKa = 3.78VTIEE375 pKa = 3.78AYY377 pKa = 9.64EE378 pKa = 4.01YY379 pKa = 10.94NEE381 pKa = 4.1SSFVGVTYY389 pKa = 10.93VSGNTYY395 pKa = 11.03DD396 pKa = 4.72SNTLTANGSYY406 pKa = 10.57YY407 pKa = 10.53LASSVPSFMNIDD419 pKa = 3.71DD420 pKa = 5.53FIQIEE425 pKa = 4.29GAGWYY430 pKa = 9.21RR431 pKa = 11.84VRR433 pKa = 11.84DD434 pKa = 3.5IIFYY438 pKa = 11.28NNIQTLILDD447 pKa = 3.82VLEE450 pKa = 4.56SDD452 pKa = 5.25FPISLNQTVKK462 pKa = 9.73GTSIYY467 pKa = 10.45NQLDD471 pKa = 3.66YY472 pKa = 11.28EE473 pKa = 4.51LYY475 pKa = 9.39EE476 pKa = 4.06ASYY479 pKa = 11.39DD480 pKa = 3.94LSTLNGDD487 pKa = 3.75YY488 pKa = 11.0YY489 pKa = 10.92LTLDD493 pKa = 3.74ATDD496 pKa = 4.2SEE498 pKa = 5.07FDD500 pKa = 3.61TVSYY504 pKa = 10.19RR505 pKa = 11.84SEE507 pKa = 3.94WFNVASMQYY516 pKa = 8.5NTYY519 pKa = 10.72LLQYY523 pKa = 9.93YY524 pKa = 10.19NSEE527 pKa = 4.25NNEE530 pKa = 4.01TNYY533 pKa = 11.03SNGIRR538 pKa = 11.84NKK540 pKa = 10.1FRR542 pKa = 11.84IRR544 pKa = 11.84YY545 pKa = 7.38QKK547 pKa = 10.33QFTYY551 pKa = 10.4IPNDD555 pKa = 3.47TNEE558 pKa = 4.12VYY560 pKa = 9.5LTDD563 pKa = 3.75TNAVQTDD570 pKa = 3.55SQYY573 pKa = 11.38RR574 pKa = 11.84DD575 pKa = 3.69YY576 pKa = 11.66LKK578 pKa = 10.74LDD580 pKa = 4.15LMPMPVNMVRR590 pKa = 11.84KK591 pKa = 10.15LGLAFSNDD599 pKa = 2.74RR600 pKa = 11.84VFINGLSLIKK610 pKa = 10.82NNEE613 pKa = 4.05LEE615 pKa = 4.06VEE617 pKa = 4.53RR618 pKa = 11.84IGEE621 pKa = 4.16SNEE624 pKa = 3.66YY625 pKa = 10.03RR626 pKa = 11.84FSAQFVRR633 pKa = 11.84SDD635 pKa = 3.31YY636 pKa = 11.53AFDD639 pKa = 3.54VTISDD644 pKa = 3.7NSIVLPQGQPLQINDD659 pKa = 3.52NAEE662 pKa = 3.74GLLFINN668 pKa = 5.04
Molecular weight: 76.04 kDa
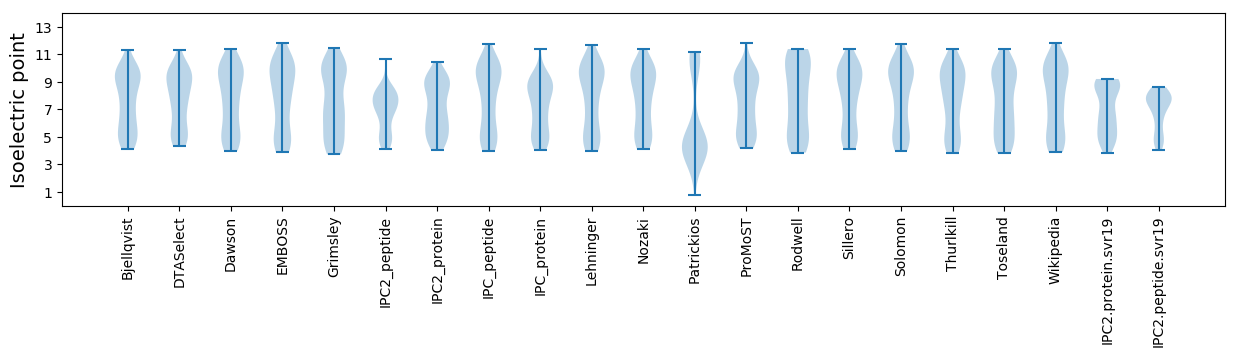
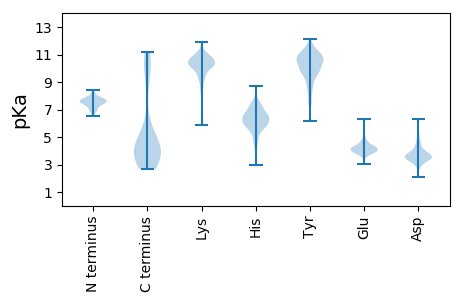
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G6XUY7|A0A6G6XUY7_9CAUD Uncharacterized protein OS=Tenacibaculum phage JQ OX=2712935 PE=4 SV=1
MM1 pKa = 7.6RR2 pKa = 11.84LIVVNRR8 pKa = 11.84RR9 pKa = 11.84YY10 pKa = 10.46LLRR13 pKa = 11.84GKK15 pKa = 9.5FFRR18 pKa = 11.84YY19 pKa = 9.96NRR21 pKa = 11.84MKK23 pKa = 10.58GVKK26 pKa = 9.97DD27 pKa = 3.67LGFKK31 pKa = 10.11FAIGIYY37 pKa = 7.61EE38 pKa = 3.77FRR40 pKa = 11.84FYY42 pKa = 11.21RR43 pKa = 4.1
MM1 pKa = 7.6RR2 pKa = 11.84LIVVNRR8 pKa = 11.84RR9 pKa = 11.84YY10 pKa = 10.46LLRR13 pKa = 11.84GKK15 pKa = 9.5FFRR18 pKa = 11.84YY19 pKa = 9.96NRR21 pKa = 11.84MKK23 pKa = 10.58GVKK26 pKa = 9.97DD27 pKa = 3.67LGFKK31 pKa = 10.11FAIGIYY37 pKa = 7.61EE38 pKa = 3.77FRR40 pKa = 11.84FYY42 pKa = 11.21RR43 pKa = 4.1
Molecular weight: 5.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11129 |
42 |
755 |
154.6 |
17.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.502 ± 0.334 | 1.339 ± 0.201 |
6.092 ± 0.208 | 7.674 ± 0.33 |
4.421 ± 0.292 | 4.996 ± 0.315 |
1.465 ± 0.199 | 7.898 ± 0.275 |
10.154 ± 0.871 | 8.536 ± 0.318 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.13 ± 0.161 | 7.79 ± 0.356 |
2.309 ± 0.151 | 3.271 ± 0.195 |
4.035 ± 0.272 | 5.895 ± 0.387 |
5.661 ± 0.314 | 5.571 ± 0.269 |
1.195 ± 0.12 | 5.068 ± 0.256 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
