
Dorea sp. CAG:317
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Dorea; environmental samples
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
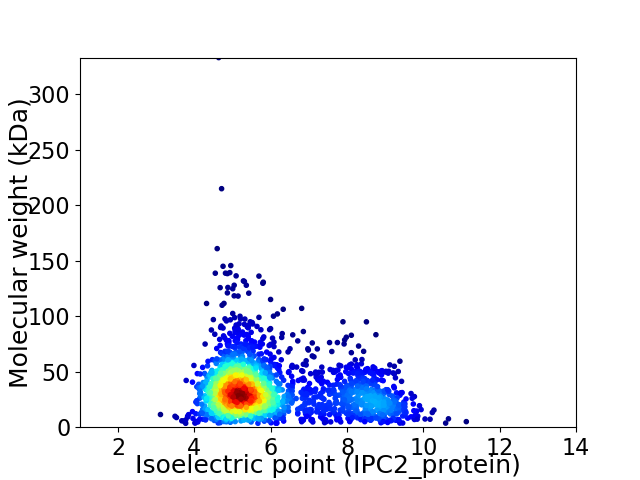
Virtual 2D-PAGE plot for 2206 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
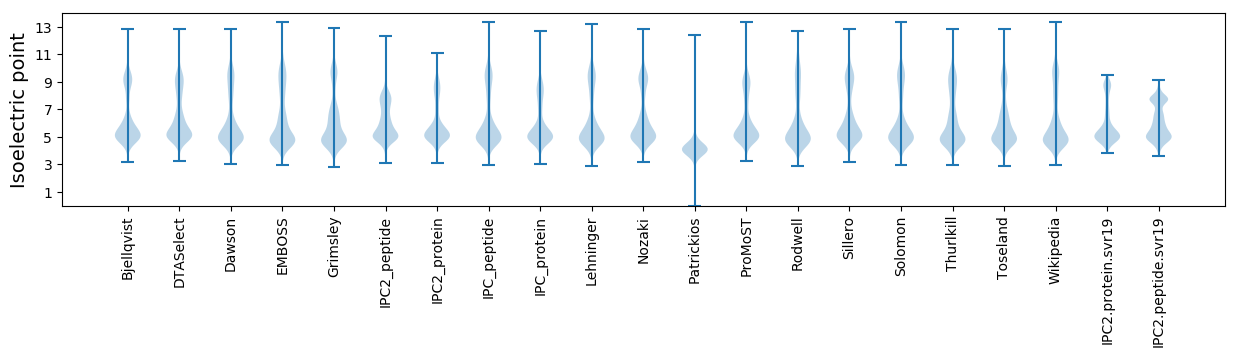
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6XNN9|R6XNN9_9FIRM 50S ribosomal protein L4 OS=Dorea sp. CAG:317 OX=1262873 GN=rplD PE=3 SV=1
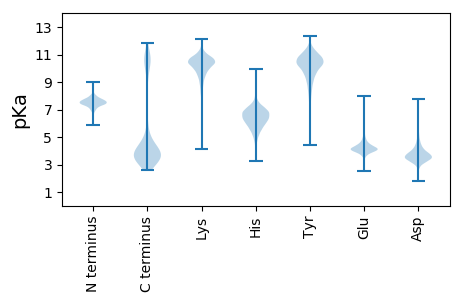
MM1 pKa = 7.82KK2 pKa = 10.34KK3 pKa = 10.11RR4 pKa = 11.84LLSVLLCAGLIASLAVGCGGGKK26 pKa = 9.87DD27 pKa = 3.29AGKK30 pKa = 10.35DD31 pKa = 3.7GEE33 pKa = 4.62KK34 pKa = 9.44EE35 pKa = 4.28TLSVWLPPLDD45 pKa = 5.46DD46 pKa = 3.89EE47 pKa = 4.85TEE49 pKa = 4.08KK50 pKa = 11.33NYY52 pKa = 10.22VPLLDD57 pKa = 4.79KK58 pKa = 11.22FEE60 pKa = 4.61EE61 pKa = 4.39EE62 pKa = 4.46NNCEE66 pKa = 4.45LDD68 pKa = 3.74VQIIPWDD75 pKa = 3.85TYY77 pKa = 8.08EE78 pKa = 4.13EE79 pKa = 4.83KK80 pKa = 11.3YY81 pKa = 7.38MTGINADD88 pKa = 3.62EE89 pKa = 4.99GPDD92 pKa = 3.38VGYY95 pKa = 9.25MYY97 pKa = 11.51VEE99 pKa = 4.26MFPTYY104 pKa = 9.21IDD106 pKa = 3.25SGAVVDD112 pKa = 3.96MSEE115 pKa = 4.09YY116 pKa = 10.95LSDD119 pKa = 3.66EE120 pKa = 4.51DD121 pKa = 4.15YY122 pKa = 11.7EE123 pKa = 4.46EE124 pKa = 4.25YY125 pKa = 10.83LYY127 pKa = 11.07LDD129 pKa = 4.18KK130 pKa = 11.68GEE132 pKa = 4.3MMGGQYY138 pKa = 10.33GVPIVTGNPFIMYY151 pKa = 9.11YY152 pKa = 10.57NKK154 pKa = 10.61DD155 pKa = 3.23ILDD158 pKa = 3.93EE159 pKa = 4.69LGEE162 pKa = 4.29KK163 pKa = 10.51APEE166 pKa = 3.61TWEE169 pKa = 3.89DD170 pKa = 3.63FKK172 pKa = 11.39RR173 pKa = 11.84ICEE176 pKa = 4.1KK177 pKa = 10.1ATKK180 pKa = 9.36DD181 pKa = 3.1TDD183 pKa = 3.62GDD185 pKa = 4.12GKK187 pKa = 10.26IDD189 pKa = 3.5QYY191 pKa = 11.91GYY193 pKa = 11.25AAGFNNGDD201 pKa = 3.58MSPLYY206 pKa = 10.56LLNSYY211 pKa = 10.29YY212 pKa = 10.89YY213 pKa = 10.76SLLWQSGSDD222 pKa = 4.1IYY224 pKa = 11.57NDD226 pKa = 3.55DD227 pKa = 3.89LKK229 pKa = 11.14SVRR232 pKa = 11.84FNDD235 pKa = 3.45EE236 pKa = 3.95AGVKK240 pKa = 9.59AVEE243 pKa = 4.2YY244 pKa = 10.22LKK246 pKa = 11.05SLTPYY251 pKa = 9.71MPEE254 pKa = 4.72DD255 pKa = 4.21YY256 pKa = 9.92MSLAATDD263 pKa = 3.82AFSSVFGGGKK273 pKa = 9.75AAFACARR280 pKa = 11.84AMQAQTEE287 pKa = 4.76SFKK290 pKa = 10.39EE291 pKa = 4.34TYY293 pKa = 10.08PDD295 pKa = 4.15LNWDD299 pKa = 3.71YY300 pKa = 10.35VTSLKK305 pKa = 10.83GEE307 pKa = 4.0QYY309 pKa = 9.87GTFGAADD316 pKa = 4.37CLTLMSACEE325 pKa = 3.92NKK327 pKa = 10.06EE328 pKa = 3.93LGMKK332 pKa = 9.9LIKK335 pKa = 10.8YY336 pKa = 7.36MVGSEE341 pKa = 4.44VMTAYY346 pKa = 10.4HH347 pKa = 6.68KK348 pKa = 10.48EE349 pKa = 4.03HH350 pKa = 6.88FGAPMTADD358 pKa = 3.24EE359 pKa = 5.43PYY361 pKa = 10.67QGDD364 pKa = 3.72EE365 pKa = 3.84KK366 pKa = 11.23LEE368 pKa = 4.67RR369 pKa = 11.84ILTEE373 pKa = 5.86DD374 pKa = 3.39RR375 pKa = 11.84DD376 pKa = 3.43KK377 pKa = 10.95WRR379 pKa = 11.84PLQAGPCGSDD389 pKa = 2.48ILLNLTSQLQAVMSGDD405 pKa = 3.81AEE407 pKa = 4.16AKK409 pKa = 8.99EE410 pKa = 4.39ALDD413 pKa = 3.79EE414 pKa = 4.38AADD417 pKa = 3.91YY418 pKa = 11.53SNEE421 pKa = 4.04LLDD424 pKa = 5.26EE425 pKa = 4.14YY426 pKa = 10.53WADD429 pKa = 3.72KK430 pKa = 10.38EE431 pKa = 4.21
MM1 pKa = 7.82KK2 pKa = 10.34KK3 pKa = 10.11RR4 pKa = 11.84LLSVLLCAGLIASLAVGCGGGKK26 pKa = 9.87DD27 pKa = 3.29AGKK30 pKa = 10.35DD31 pKa = 3.7GEE33 pKa = 4.62KK34 pKa = 9.44EE35 pKa = 4.28TLSVWLPPLDD45 pKa = 5.46DD46 pKa = 3.89EE47 pKa = 4.85TEE49 pKa = 4.08KK50 pKa = 11.33NYY52 pKa = 10.22VPLLDD57 pKa = 4.79KK58 pKa = 11.22FEE60 pKa = 4.61EE61 pKa = 4.39EE62 pKa = 4.46NNCEE66 pKa = 4.45LDD68 pKa = 3.74VQIIPWDD75 pKa = 3.85TYY77 pKa = 8.08EE78 pKa = 4.13EE79 pKa = 4.83KK80 pKa = 11.3YY81 pKa = 7.38MTGINADD88 pKa = 3.62EE89 pKa = 4.99GPDD92 pKa = 3.38VGYY95 pKa = 9.25MYY97 pKa = 11.51VEE99 pKa = 4.26MFPTYY104 pKa = 9.21IDD106 pKa = 3.25SGAVVDD112 pKa = 3.96MSEE115 pKa = 4.09YY116 pKa = 10.95LSDD119 pKa = 3.66EE120 pKa = 4.51DD121 pKa = 4.15YY122 pKa = 11.7EE123 pKa = 4.46EE124 pKa = 4.25YY125 pKa = 10.83LYY127 pKa = 11.07LDD129 pKa = 4.18KK130 pKa = 11.68GEE132 pKa = 4.3MMGGQYY138 pKa = 10.33GVPIVTGNPFIMYY151 pKa = 9.11YY152 pKa = 10.57NKK154 pKa = 10.61DD155 pKa = 3.23ILDD158 pKa = 3.93EE159 pKa = 4.69LGEE162 pKa = 4.29KK163 pKa = 10.51APEE166 pKa = 3.61TWEE169 pKa = 3.89DD170 pKa = 3.63FKK172 pKa = 11.39RR173 pKa = 11.84ICEE176 pKa = 4.1KK177 pKa = 10.1ATKK180 pKa = 9.36DD181 pKa = 3.1TDD183 pKa = 3.62GDD185 pKa = 4.12GKK187 pKa = 10.26IDD189 pKa = 3.5QYY191 pKa = 11.91GYY193 pKa = 11.25AAGFNNGDD201 pKa = 3.58MSPLYY206 pKa = 10.56LLNSYY211 pKa = 10.29YY212 pKa = 10.89YY213 pKa = 10.76SLLWQSGSDD222 pKa = 4.1IYY224 pKa = 11.57NDD226 pKa = 3.55DD227 pKa = 3.89LKK229 pKa = 11.14SVRR232 pKa = 11.84FNDD235 pKa = 3.45EE236 pKa = 3.95AGVKK240 pKa = 9.59AVEE243 pKa = 4.2YY244 pKa = 10.22LKK246 pKa = 11.05SLTPYY251 pKa = 9.71MPEE254 pKa = 4.72DD255 pKa = 4.21YY256 pKa = 9.92MSLAATDD263 pKa = 3.82AFSSVFGGGKK273 pKa = 9.75AAFACARR280 pKa = 11.84AMQAQTEE287 pKa = 4.76SFKK290 pKa = 10.39EE291 pKa = 4.34TYY293 pKa = 10.08PDD295 pKa = 4.15LNWDD299 pKa = 3.71YY300 pKa = 10.35VTSLKK305 pKa = 10.83GEE307 pKa = 4.0QYY309 pKa = 9.87GTFGAADD316 pKa = 4.37CLTLMSACEE325 pKa = 3.92NKK327 pKa = 10.06EE328 pKa = 3.93LGMKK332 pKa = 9.9LIKK335 pKa = 10.8YY336 pKa = 7.36MVGSEE341 pKa = 4.44VMTAYY346 pKa = 10.4HH347 pKa = 6.68KK348 pKa = 10.48EE349 pKa = 4.03HH350 pKa = 6.88FGAPMTADD358 pKa = 3.24EE359 pKa = 5.43PYY361 pKa = 10.67QGDD364 pKa = 3.72EE365 pKa = 3.84KK366 pKa = 11.23LEE368 pKa = 4.67RR369 pKa = 11.84ILTEE373 pKa = 5.86DD374 pKa = 3.39RR375 pKa = 11.84DD376 pKa = 3.43KK377 pKa = 10.95WRR379 pKa = 11.84PLQAGPCGSDD389 pKa = 2.48ILLNLTSQLQAVMSGDD405 pKa = 3.81AEE407 pKa = 4.16AKK409 pKa = 8.99EE410 pKa = 4.39ALDD413 pKa = 3.79EE414 pKa = 4.38AADD417 pKa = 3.91YY418 pKa = 11.53SNEE421 pKa = 4.04LLDD424 pKa = 5.26EE425 pKa = 4.14YY426 pKa = 10.53WADD429 pKa = 3.72KK430 pKa = 10.38EE431 pKa = 4.21
Molecular weight: 48.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6XJZ0|R6XJZ0_9FIRM Tripeptide aminopeptidase OS=Dorea sp. CAG:317 OX=1262873 GN=BN605_02204 PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.79KK9 pKa = 7.6RR10 pKa = 11.84QRR12 pKa = 11.84AKK14 pKa = 9.2VHH16 pKa = 5.65GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.71VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 8.83KK41 pKa = 10.63LSAA44 pKa = 3.95
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.79KK9 pKa = 7.6RR10 pKa = 11.84QRR12 pKa = 11.84AKK14 pKa = 9.2VHH16 pKa = 5.65GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.71VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 8.83KK41 pKa = 10.63LSAA44 pKa = 3.95
Molecular weight: 5.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
683009 |
29 |
3000 |
309.6 |
34.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.377 ± 0.047 | 1.48 ± 0.023 |
5.472 ± 0.043 | 8.31 ± 0.071 |
3.975 ± 0.038 | 7.249 ± 0.048 |
1.785 ± 0.021 | 7.387 ± 0.049 |
7.25 ± 0.042 | 8.976 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.243 ± 0.03 | 4.08 ± 0.036 |
3.151 ± 0.024 | 3.156 ± 0.03 |
4.29 ± 0.038 | 5.414 ± 0.038 |
5.277 ± 0.046 | 7.221 ± 0.043 |
0.82 ± 0.019 | 4.085 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
