
Torque teno midi virus 2
Taxonomy: Viruses; Anelloviridae; Gammatorquevirus
Average proteome isoelectric point is 7.45
Get precalculated fractions of proteins
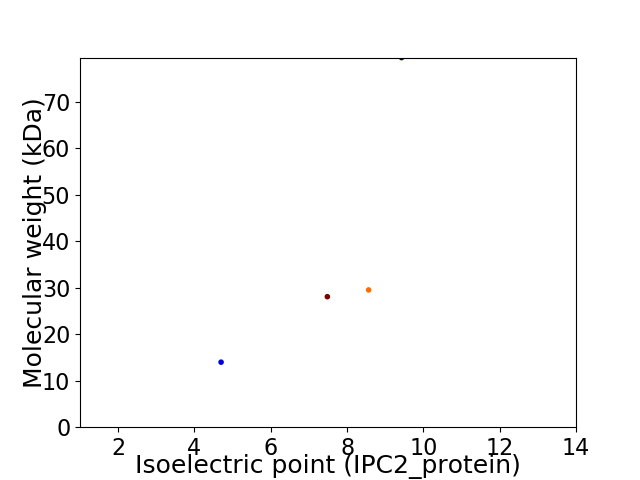
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
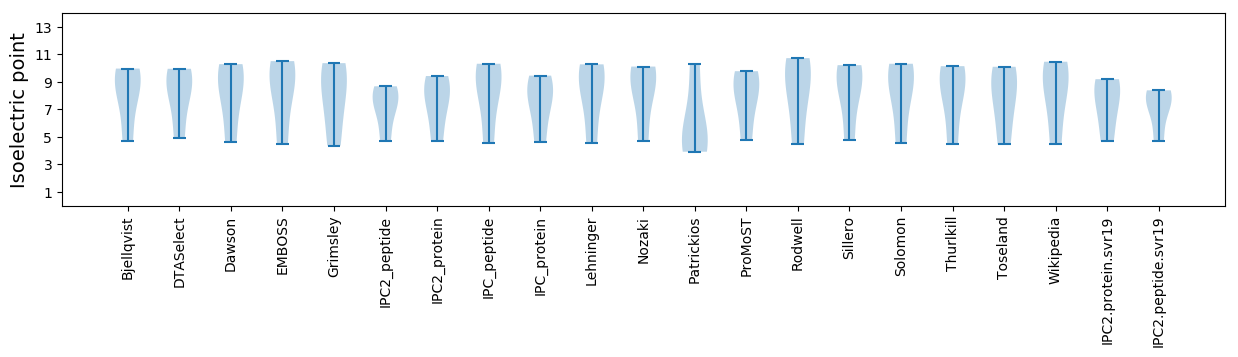
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A4GZA1|A4GZA1_9VIRU Capsid protein OS=Torque teno midi virus 2 OX=687380 PE=3 SV=1
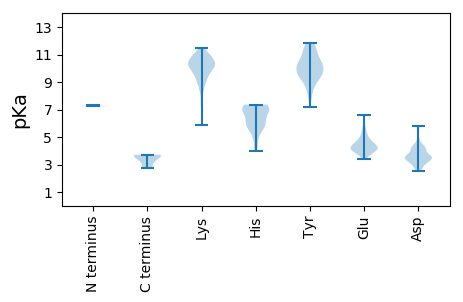
MM1 pKa = 7.24QNISYY6 pKa = 10.24TDD8 pKa = 4.05FYY10 pKa = 11.48KK11 pKa = 9.67PTPYY15 pKa = 9.58NTDD18 pKa = 2.73TKK20 pKa = 9.85NQIWLSQLADD30 pKa = 4.2SHH32 pKa = 7.36DD33 pKa = 3.57NWCNCDD39 pKa = 3.79FPFAHH44 pKa = 7.26LLASIFPPGHH54 pKa = 6.98KK55 pKa = 10.31DD56 pKa = 2.73RR57 pKa = 11.84TRR59 pKa = 11.84TINQILQRR67 pKa = 11.84DD68 pKa = 4.31FRR70 pKa = 11.84QKK72 pKa = 10.5CLSGGADD79 pKa = 3.34AKK81 pKa = 11.19DD82 pKa = 3.29SGLAGEE88 pKa = 5.2DD89 pKa = 4.13LEE91 pKa = 6.61KK92 pKa = 10.99DD93 pKa = 3.61GQKK96 pKa = 10.98DD97 pKa = 3.15GGKK100 pKa = 8.49YY101 pKa = 8.52TEE103 pKa = 4.52EE104 pKa = 4.18DD105 pKa = 3.47LPRR108 pKa = 11.84DD109 pKa = 3.52EE110 pKa = 5.63VEE112 pKa = 4.12EE113 pKa = 3.85LLAAVEE119 pKa = 4.64SAASRR124 pKa = 3.62
MM1 pKa = 7.24QNISYY6 pKa = 10.24TDD8 pKa = 4.05FYY10 pKa = 11.48KK11 pKa = 9.67PTPYY15 pKa = 9.58NTDD18 pKa = 2.73TKK20 pKa = 9.85NQIWLSQLADD30 pKa = 4.2SHH32 pKa = 7.36DD33 pKa = 3.57NWCNCDD39 pKa = 3.79FPFAHH44 pKa = 7.26LLASIFPPGHH54 pKa = 6.98KK55 pKa = 10.31DD56 pKa = 2.73RR57 pKa = 11.84TRR59 pKa = 11.84TINQILQRR67 pKa = 11.84DD68 pKa = 4.31FRR70 pKa = 11.84QKK72 pKa = 10.5CLSGGADD79 pKa = 3.34AKK81 pKa = 11.19DD82 pKa = 3.29SGLAGEE88 pKa = 5.2DD89 pKa = 4.13LEE91 pKa = 6.61KK92 pKa = 10.99DD93 pKa = 3.61GQKK96 pKa = 10.98DD97 pKa = 3.15GGKK100 pKa = 8.49YY101 pKa = 8.52TEE103 pKa = 4.52EE104 pKa = 4.18DD105 pKa = 3.47LPRR108 pKa = 11.84DD109 pKa = 3.52EE110 pKa = 5.63VEE112 pKa = 4.12EE113 pKa = 3.85LLAAVEE119 pKa = 4.64SAASRR124 pKa = 3.62
Molecular weight: 13.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A4GZA1|A4GZA1_9VIRU Capsid protein OS=Torque teno midi virus 2 OX=687380 PE=3 SV=1
MM1 pKa = 7.35PFWWGRR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 10.22RR10 pKa = 11.84FWLGRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84FRR19 pKa = 11.84KK20 pKa = 8.78RR21 pKa = 11.84WPKK24 pKa = 9.9RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84KK28 pKa = 7.51IHH30 pKa = 6.02RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84FTKK36 pKa = 10.08RR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84RR41 pKa = 11.84TTRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84KK48 pKa = 9.21RR49 pKa = 11.84RR50 pKa = 11.84FKK52 pKa = 10.79VRR54 pKa = 11.84RR55 pKa = 11.84KK56 pKa = 9.51KK57 pKa = 10.47PKK59 pKa = 9.15IPIYY63 pKa = 9.73QWQPDD68 pKa = 4.33SIRR71 pKa = 11.84KK72 pKa = 9.16CKK74 pKa = 10.08IKK76 pKa = 10.8GVGTLVLGAHH86 pKa = 7.12GKK88 pKa = 9.59QFVCYY93 pKa = 9.31TDD95 pKa = 3.25VEE97 pKa = 4.81TRR99 pKa = 11.84APPPKK104 pKa = 10.39APGGGGFGCKK114 pKa = 8.44QFSLQYY120 pKa = 10.42LYY122 pKa = 11.14EE123 pKa = 4.19EE124 pKa = 4.13YY125 pKa = 10.63RR126 pKa = 11.84FRR128 pKa = 11.84NNIWTHH134 pKa = 4.39TNINLDD140 pKa = 3.4LVRR143 pKa = 11.84YY144 pKa = 9.23LRR146 pKa = 11.84AAFTFYY152 pKa = 10.65RR153 pKa = 11.84HH154 pKa = 6.56PDD156 pKa = 2.51IDD158 pKa = 4.5FIINYY163 pKa = 8.94DD164 pKa = 3.87RR165 pKa = 11.84QPPFYY170 pKa = 9.82LDD172 pKa = 2.95KK173 pKa = 10.33FTYY176 pKa = 9.35PLCHH180 pKa = 6.4PQNLLLGKK188 pKa = 9.87HH189 pKa = 6.05KK190 pKa = 10.53IILLSKK196 pKa = 10.09ASKK199 pKa = 10.57PNGKK203 pKa = 8.95VKK205 pKa = 10.38KK206 pKa = 10.1RR207 pKa = 11.84IIIKK211 pKa = 9.68PPKK214 pKa = 10.17QMITKK219 pKa = 9.29WFFQEE224 pKa = 4.3QFTTQPLLSLRR235 pKa = 11.84AAAASFQYY243 pKa = 10.07PHH245 pKa = 7.26IGCCMPNRR253 pKa = 11.84VVTFSALNPGFYY265 pKa = 9.96QQGNWSLAQQEE276 pKa = 4.41THH278 pKa = 7.24PYY280 pKa = 9.62SPWPQIPRR288 pKa = 11.84RR289 pKa = 11.84LIFWDD294 pKa = 4.25VPASEE299 pKa = 5.15IPTDD303 pKa = 3.65GQQRR307 pKa = 11.84LDD309 pKa = 3.62FSKK312 pKa = 10.44KK313 pKa = 9.27HH314 pKa = 6.0AFITDD319 pKa = 2.9IHH321 pKa = 7.14SYY323 pKa = 10.26SDD325 pKa = 3.3SVSYY329 pKa = 11.04NKK331 pKa = 10.6GYY333 pKa = 7.92FTSKK337 pKa = 9.62ILKK340 pKa = 10.07AKK342 pKa = 9.93FVSKK346 pKa = 10.49DD347 pKa = 3.05ISDD350 pKa = 3.57TAGIGNLPLNLCRR363 pKa = 11.84YY364 pKa = 9.41NPAIDD369 pKa = 3.47TGKK372 pKa = 11.15GNTIWLHH379 pKa = 5.47SNLVNSYY386 pKa = 10.82SKK388 pKa = 10.02PQHH391 pKa = 6.82DD392 pKa = 4.37EE393 pKa = 4.51DD394 pKa = 5.81IIITGMPIWMLLFGWLSYY412 pKa = 9.29VQHH415 pKa = 6.05VKK417 pKa = 10.58KK418 pKa = 10.54PPDD421 pKa = 3.78FYY423 pKa = 11.6LSFTVLIEE431 pKa = 4.17SPAIEE436 pKa = 4.31VASASATPTPVIPIDD451 pKa = 3.38TSFINGNPPYY461 pKa = 9.97NQTITIRR468 pKa = 11.84DD469 pKa = 3.72KK470 pKa = 10.86AHH472 pKa = 6.61WWPDD476 pKa = 3.13VYY478 pKa = 11.49NQTEE482 pKa = 4.3VLNSLVTAGPYY493 pKa = 9.33VPKK496 pKa = 10.35LDD498 pKa = 4.49NIRR501 pKa = 11.84NSTWEE506 pKa = 3.39LHH508 pKa = 5.22YY509 pKa = 10.39FYY511 pKa = 11.31NFLFKK516 pKa = 10.25WGGPEE521 pKa = 4.17STDD524 pKa = 3.6QPVADD529 pKa = 4.87PSKK532 pKa = 10.81QPIYY536 pKa = 10.62DD537 pKa = 4.08ALDD540 pKa = 3.56KK541 pKa = 10.79QQQTVQIRR549 pKa = 11.84NPAKK553 pKa = 10.04QKK555 pKa = 10.26YY556 pKa = 7.21ATIMHH561 pKa = 6.99PWDD564 pKa = 3.42VRR566 pKa = 11.84RR567 pKa = 11.84GIITNKK573 pKa = 9.42ALKK576 pKa = 10.28RR577 pKa = 11.84MYY579 pKa = 10.2EE580 pKa = 3.98NLSIDD585 pKa = 3.6STFEE589 pKa = 3.92ADD591 pKa = 3.95SIASKK596 pKa = 10.34RR597 pKa = 11.84KK598 pKa = 9.41RR599 pKa = 11.84VTGPCFTALQEE610 pKa = 4.23INQEE614 pKa = 3.93EE615 pKa = 4.71RR616 pKa = 11.84TCLQTLCEE624 pKa = 4.0EE625 pKa = 4.89NIFQEE630 pKa = 4.8TPQEE634 pKa = 4.0EE635 pKa = 4.37NLQQLILQQQQHH647 pKa = 4.92QQDD650 pKa = 3.12IKK652 pKa = 11.17YY653 pKa = 10.33NILKK657 pKa = 10.27LISQMKK663 pKa = 8.29EE664 pKa = 3.57QQNMLKK670 pKa = 10.46LHH672 pKa = 6.43TGLLNN677 pKa = 3.66
MM1 pKa = 7.35PFWWGRR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 10.22RR10 pKa = 11.84FWLGRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84FRR19 pKa = 11.84KK20 pKa = 8.78RR21 pKa = 11.84WPKK24 pKa = 9.9RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84KK28 pKa = 7.51IHH30 pKa = 6.02RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84FTKK36 pKa = 10.08RR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84RR41 pKa = 11.84TTRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84KK48 pKa = 9.21RR49 pKa = 11.84RR50 pKa = 11.84FKK52 pKa = 10.79VRR54 pKa = 11.84RR55 pKa = 11.84KK56 pKa = 9.51KK57 pKa = 10.47PKK59 pKa = 9.15IPIYY63 pKa = 9.73QWQPDD68 pKa = 4.33SIRR71 pKa = 11.84KK72 pKa = 9.16CKK74 pKa = 10.08IKK76 pKa = 10.8GVGTLVLGAHH86 pKa = 7.12GKK88 pKa = 9.59QFVCYY93 pKa = 9.31TDD95 pKa = 3.25VEE97 pKa = 4.81TRR99 pKa = 11.84APPPKK104 pKa = 10.39APGGGGFGCKK114 pKa = 8.44QFSLQYY120 pKa = 10.42LYY122 pKa = 11.14EE123 pKa = 4.19EE124 pKa = 4.13YY125 pKa = 10.63RR126 pKa = 11.84FRR128 pKa = 11.84NNIWTHH134 pKa = 4.39TNINLDD140 pKa = 3.4LVRR143 pKa = 11.84YY144 pKa = 9.23LRR146 pKa = 11.84AAFTFYY152 pKa = 10.65RR153 pKa = 11.84HH154 pKa = 6.56PDD156 pKa = 2.51IDD158 pKa = 4.5FIINYY163 pKa = 8.94DD164 pKa = 3.87RR165 pKa = 11.84QPPFYY170 pKa = 9.82LDD172 pKa = 2.95KK173 pKa = 10.33FTYY176 pKa = 9.35PLCHH180 pKa = 6.4PQNLLLGKK188 pKa = 9.87HH189 pKa = 6.05KK190 pKa = 10.53IILLSKK196 pKa = 10.09ASKK199 pKa = 10.57PNGKK203 pKa = 8.95VKK205 pKa = 10.38KK206 pKa = 10.1RR207 pKa = 11.84IIIKK211 pKa = 9.68PPKK214 pKa = 10.17QMITKK219 pKa = 9.29WFFQEE224 pKa = 4.3QFTTQPLLSLRR235 pKa = 11.84AAAASFQYY243 pKa = 10.07PHH245 pKa = 7.26IGCCMPNRR253 pKa = 11.84VVTFSALNPGFYY265 pKa = 9.96QQGNWSLAQQEE276 pKa = 4.41THH278 pKa = 7.24PYY280 pKa = 9.62SPWPQIPRR288 pKa = 11.84RR289 pKa = 11.84LIFWDD294 pKa = 4.25VPASEE299 pKa = 5.15IPTDD303 pKa = 3.65GQQRR307 pKa = 11.84LDD309 pKa = 3.62FSKK312 pKa = 10.44KK313 pKa = 9.27HH314 pKa = 6.0AFITDD319 pKa = 2.9IHH321 pKa = 7.14SYY323 pKa = 10.26SDD325 pKa = 3.3SVSYY329 pKa = 11.04NKK331 pKa = 10.6GYY333 pKa = 7.92FTSKK337 pKa = 9.62ILKK340 pKa = 10.07AKK342 pKa = 9.93FVSKK346 pKa = 10.49DD347 pKa = 3.05ISDD350 pKa = 3.57TAGIGNLPLNLCRR363 pKa = 11.84YY364 pKa = 9.41NPAIDD369 pKa = 3.47TGKK372 pKa = 11.15GNTIWLHH379 pKa = 5.47SNLVNSYY386 pKa = 10.82SKK388 pKa = 10.02PQHH391 pKa = 6.82DD392 pKa = 4.37EE393 pKa = 4.51DD394 pKa = 5.81IIITGMPIWMLLFGWLSYY412 pKa = 9.29VQHH415 pKa = 6.05VKK417 pKa = 10.58KK418 pKa = 10.54PPDD421 pKa = 3.78FYY423 pKa = 11.6LSFTVLIEE431 pKa = 4.17SPAIEE436 pKa = 4.31VASASATPTPVIPIDD451 pKa = 3.38TSFINGNPPYY461 pKa = 9.97NQTITIRR468 pKa = 11.84DD469 pKa = 3.72KK470 pKa = 10.86AHH472 pKa = 6.61WWPDD476 pKa = 3.13VYY478 pKa = 11.49NQTEE482 pKa = 4.3VLNSLVTAGPYY493 pKa = 9.33VPKK496 pKa = 10.35LDD498 pKa = 4.49NIRR501 pKa = 11.84NSTWEE506 pKa = 3.39LHH508 pKa = 5.22YY509 pKa = 10.39FYY511 pKa = 11.31NFLFKK516 pKa = 10.25WGGPEE521 pKa = 4.17STDD524 pKa = 3.6QPVADD529 pKa = 4.87PSKK532 pKa = 10.81QPIYY536 pKa = 10.62DD537 pKa = 4.08ALDD540 pKa = 3.56KK541 pKa = 10.79QQQTVQIRR549 pKa = 11.84NPAKK553 pKa = 10.04QKK555 pKa = 10.26YY556 pKa = 7.21ATIMHH561 pKa = 6.99PWDD564 pKa = 3.42VRR566 pKa = 11.84RR567 pKa = 11.84GIITNKK573 pKa = 9.42ALKK576 pKa = 10.28RR577 pKa = 11.84MYY579 pKa = 10.2EE580 pKa = 3.98NLSIDD585 pKa = 3.6STFEE589 pKa = 3.92ADD591 pKa = 3.95SIASKK596 pKa = 10.34RR597 pKa = 11.84KK598 pKa = 9.41RR599 pKa = 11.84VTGPCFTALQEE610 pKa = 4.23INQEE614 pKa = 3.93EE615 pKa = 4.71RR616 pKa = 11.84TCLQTLCEE624 pKa = 4.0EE625 pKa = 4.89NIFQEE630 pKa = 4.8TPQEE634 pKa = 4.0EE635 pKa = 4.37NLQQLILQQQQHH647 pKa = 4.92QQDD650 pKa = 3.12IKK652 pKa = 11.17YY653 pKa = 10.33NILKK657 pKa = 10.27LISQMKK663 pKa = 8.29EE664 pKa = 3.57QQNMLKK670 pKa = 10.46LHH672 pKa = 6.43TGLLNN677 pKa = 3.66
Molecular weight: 79.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1310 |
124 |
677 |
327.5 |
37.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.725 ± 0.906 | 1.603 ± 0.171 |
5.802 ± 1.275 | 4.58 ± 0.862 |
4.427 ± 0.585 | 4.885 ± 0.3 |
2.748 ± 0.209 | 5.878 ± 1.201 |
8.015 ± 0.556 | 8.092 ± 0.287 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.45 ± 0.2 | 5.344 ± 0.29 |
7.023 ± 0.674 | 6.489 ± 0.577 |
6.489 ± 0.63 | 6.794 ± 1.265 |
6.412 ± 0.302 | 2.824 ± 0.645 |
1.908 ± 0.458 | 3.511 ± 0.487 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
