
Changping earthworm virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
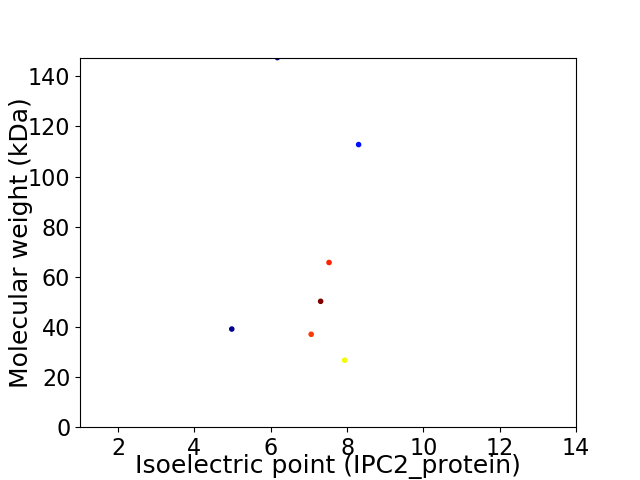
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KKP1|A0A1L3KKP1_9VIRU Uncharacterized protein OS=Changping earthworm virus 2 OX=1922827 PE=4 SV=1
MM1 pKa = 7.52CSDD4 pKa = 3.01VSGRR8 pKa = 11.84LSVEE12 pKa = 3.74LAKK15 pKa = 10.52CRR17 pKa = 11.84QVSFPKK23 pKa = 10.66GVIVTLGPKK32 pKa = 9.93CDD34 pKa = 3.91GSDD37 pKa = 3.54CYY39 pKa = 11.72GEE41 pKa = 4.38FFPLITFTEE50 pKa = 4.23KK51 pKa = 10.62CFPEE55 pKa = 5.69FIRR58 pKa = 11.84DD59 pKa = 3.44WGWALFNVVCNCTTASTPLSISRR82 pKa = 11.84EE83 pKa = 3.91EE84 pKa = 4.09SAKK87 pKa = 10.65SCFISKK93 pKa = 8.35STFLEE98 pKa = 5.11GYY100 pKa = 8.95GRR102 pKa = 11.84YY103 pKa = 9.55GADD106 pKa = 3.44PGWAARR112 pKa = 11.84TLVRR116 pKa = 11.84PKK118 pKa = 9.51FTTTIEE124 pKa = 4.17EE125 pKa = 3.97FEE127 pKa = 4.32EE128 pKa = 4.85WVGHH132 pKa = 5.67IANVAAGNRR141 pKa = 11.84GPAGSQEE148 pKa = 4.0SGEE151 pKa = 4.07AHH153 pKa = 6.98FSNVDD158 pKa = 3.02QWGGRR163 pKa = 11.84IAGTDD168 pKa = 3.5FVVRR172 pKa = 11.84HH173 pKa = 4.9VTSEE177 pKa = 4.22CEE179 pKa = 3.93LEE181 pKa = 4.03RR182 pKa = 11.84GYY184 pKa = 11.42KK185 pKa = 9.55FAGPNKK191 pKa = 10.06DD192 pKa = 3.7LEE194 pKa = 4.09LHH196 pKa = 5.58LRR198 pKa = 11.84YY199 pKa = 9.77RR200 pKa = 11.84PFFHH204 pKa = 7.42IEE206 pKa = 3.45TSTAGTLGVRR216 pKa = 11.84VFGSPIYY223 pKa = 9.45WAMWSYY229 pKa = 10.43FAHH232 pKa = 6.29NPGITGAEE240 pKa = 3.6QHH242 pKa = 6.17IEE244 pKa = 3.94KK245 pKa = 10.57FNLKK249 pKa = 9.95FGEE252 pKa = 4.2AAWIQKK258 pKa = 10.12VGIIYY263 pKa = 10.28KK264 pKa = 10.03DD265 pKa = 3.55VPDD268 pKa = 4.46EE269 pKa = 4.75GDD271 pKa = 3.14VEE273 pKa = 4.4YY274 pKa = 10.84QGEE277 pKa = 4.21DD278 pKa = 3.28EE279 pKa = 4.47EE280 pKa = 5.54EE281 pKa = 4.24YY282 pKa = 10.98DD283 pKa = 3.83GSEE286 pKa = 4.14EE287 pKa = 4.86LEE289 pKa = 4.13EE290 pKa = 5.26LPTLSDD296 pKa = 3.26IEE298 pKa = 4.43VNPEE302 pKa = 3.62EE303 pKa = 3.95IQARR307 pKa = 11.84QEE309 pKa = 4.09SALLEE314 pKa = 4.02MSALKK319 pKa = 10.67DD320 pKa = 3.38SLPRR324 pKa = 11.84AFYY327 pKa = 10.6KK328 pKa = 10.15PLPRR332 pKa = 11.84VQRR335 pKa = 11.84SDD337 pKa = 3.06FLQRR341 pKa = 11.84YY342 pKa = 4.17TAPYY346 pKa = 9.79RR347 pKa = 11.84KK348 pKa = 9.74
MM1 pKa = 7.52CSDD4 pKa = 3.01VSGRR8 pKa = 11.84LSVEE12 pKa = 3.74LAKK15 pKa = 10.52CRR17 pKa = 11.84QVSFPKK23 pKa = 10.66GVIVTLGPKK32 pKa = 9.93CDD34 pKa = 3.91GSDD37 pKa = 3.54CYY39 pKa = 11.72GEE41 pKa = 4.38FFPLITFTEE50 pKa = 4.23KK51 pKa = 10.62CFPEE55 pKa = 5.69FIRR58 pKa = 11.84DD59 pKa = 3.44WGWALFNVVCNCTTASTPLSISRR82 pKa = 11.84EE83 pKa = 3.91EE84 pKa = 4.09SAKK87 pKa = 10.65SCFISKK93 pKa = 8.35STFLEE98 pKa = 5.11GYY100 pKa = 8.95GRR102 pKa = 11.84YY103 pKa = 9.55GADD106 pKa = 3.44PGWAARR112 pKa = 11.84TLVRR116 pKa = 11.84PKK118 pKa = 9.51FTTTIEE124 pKa = 4.17EE125 pKa = 3.97FEE127 pKa = 4.32EE128 pKa = 4.85WVGHH132 pKa = 5.67IANVAAGNRR141 pKa = 11.84GPAGSQEE148 pKa = 4.0SGEE151 pKa = 4.07AHH153 pKa = 6.98FSNVDD158 pKa = 3.02QWGGRR163 pKa = 11.84IAGTDD168 pKa = 3.5FVVRR172 pKa = 11.84HH173 pKa = 4.9VTSEE177 pKa = 4.22CEE179 pKa = 3.93LEE181 pKa = 4.03RR182 pKa = 11.84GYY184 pKa = 11.42KK185 pKa = 9.55FAGPNKK191 pKa = 10.06DD192 pKa = 3.7LEE194 pKa = 4.09LHH196 pKa = 5.58LRR198 pKa = 11.84YY199 pKa = 9.77RR200 pKa = 11.84PFFHH204 pKa = 7.42IEE206 pKa = 3.45TSTAGTLGVRR216 pKa = 11.84VFGSPIYY223 pKa = 9.45WAMWSYY229 pKa = 10.43FAHH232 pKa = 6.29NPGITGAEE240 pKa = 3.6QHH242 pKa = 6.17IEE244 pKa = 3.94KK245 pKa = 10.57FNLKK249 pKa = 9.95FGEE252 pKa = 4.2AAWIQKK258 pKa = 10.12VGIIYY263 pKa = 10.28KK264 pKa = 10.03DD265 pKa = 3.55VPDD268 pKa = 4.46EE269 pKa = 4.75GDD271 pKa = 3.14VEE273 pKa = 4.4YY274 pKa = 10.84QGEE277 pKa = 4.21DD278 pKa = 3.28EE279 pKa = 4.47EE280 pKa = 5.54EE281 pKa = 4.24YY282 pKa = 10.98DD283 pKa = 3.83GSEE286 pKa = 4.14EE287 pKa = 4.86LEE289 pKa = 4.13EE290 pKa = 5.26LPTLSDD296 pKa = 3.26IEE298 pKa = 4.43VNPEE302 pKa = 3.62EE303 pKa = 3.95IQARR307 pKa = 11.84QEE309 pKa = 4.09SALLEE314 pKa = 4.02MSALKK319 pKa = 10.67DD320 pKa = 3.38SLPRR324 pKa = 11.84AFYY327 pKa = 10.6KK328 pKa = 10.15PLPRR332 pKa = 11.84VQRR335 pKa = 11.84SDD337 pKa = 3.06FLQRR341 pKa = 11.84YY342 pKa = 4.17TAPYY346 pKa = 9.79RR347 pKa = 11.84KK348 pKa = 9.74
Molecular weight: 39.16 kDa
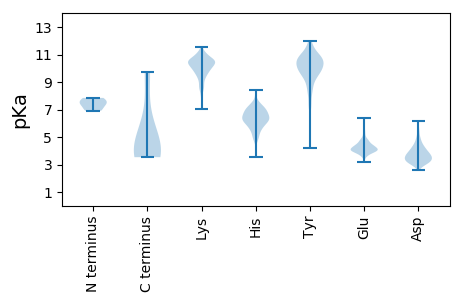
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KKH0|A0A1L3KKH0_9VIRU Uncharacterized protein OS=Changping earthworm virus 2 OX=1922827 PE=4 SV=1
MM1 pKa = 6.91NHH3 pKa = 6.22RR4 pKa = 11.84MNSRR8 pKa = 11.84KK9 pKa = 9.31QKK11 pKa = 10.43HH12 pKa = 5.5GDD14 pKa = 3.02MMNYY18 pKa = 10.41NMTNIYY24 pKa = 8.03NTNAMMRR31 pKa = 11.84IRR33 pKa = 11.84KK34 pKa = 9.19RR35 pKa = 11.84KK36 pKa = 9.63INDD39 pKa = 3.53AEE41 pKa = 4.1DD42 pKa = 3.15MRR44 pKa = 11.84AKK46 pKa = 10.36RR47 pKa = 11.84KK48 pKa = 9.9CLEE51 pKa = 4.24DD52 pKa = 3.37LRR54 pKa = 11.84KK55 pKa = 9.95LRR57 pKa = 11.84TEE59 pKa = 3.63THH61 pKa = 4.78EE62 pKa = 3.92TGFRR66 pKa = 11.84IEE68 pKa = 4.06TLRR71 pKa = 11.84AAEE74 pKa = 4.83LLMEE78 pKa = 5.03DD79 pKa = 4.62AGFSDD84 pKa = 4.02STVTRR89 pKa = 11.84ANSKK93 pKa = 9.8LLRR96 pKa = 11.84NLGHH100 pKa = 7.13KK101 pKa = 9.93IKK103 pKa = 10.44PPNIQLHH110 pKa = 6.05NISMNRR116 pKa = 11.84FSSIMVKK123 pKa = 9.53TSEE126 pKa = 4.39YY127 pKa = 10.09EE128 pKa = 4.05LNSLGIEE135 pKa = 4.59PIHH138 pKa = 6.19AAGRR142 pKa = 11.84TYY144 pKa = 10.79IKK146 pKa = 10.67FQDD149 pKa = 4.02LSKK152 pKa = 10.41LCCSRR157 pKa = 11.84TTDD160 pKa = 3.67FDD162 pKa = 3.76SALAEE167 pKa = 4.3FFNLMSQRR175 pKa = 11.84TQRR178 pKa = 11.84SYY180 pKa = 11.8NLWKK184 pKa = 10.82SLVDD188 pKa = 3.26EE189 pKa = 4.5AQILEE194 pKa = 4.38VPRR197 pKa = 11.84VPTHH201 pKa = 5.25QLQEE205 pKa = 3.78VVMRR209 pKa = 11.84DD210 pKa = 3.03KK211 pKa = 11.21VLPTTSSPLEE221 pKa = 3.82VLSYY225 pKa = 9.9CFPAVYY231 pKa = 9.93PNVVRR236 pKa = 11.84LSPLAMVEE244 pKa = 3.66NDD246 pKa = 3.08AKK248 pKa = 10.74FVEE251 pKa = 4.44NVIRR255 pKa = 11.84MVKK258 pKa = 9.94SLHH261 pKa = 6.21FKK263 pKa = 9.61MLNQIDD269 pKa = 4.45GEE271 pKa = 4.65VLGHH275 pKa = 6.83HH276 pKa = 6.55LMKK279 pKa = 11.08LMTEE283 pKa = 4.35DD284 pKa = 3.46KK285 pKa = 10.94GLRR288 pKa = 11.84NCSRR292 pKa = 11.84ATPEE296 pKa = 3.79GFEE299 pKa = 4.06MFSCFFNNTYY309 pKa = 9.72EE310 pKa = 4.16SHH312 pKa = 6.69SISINMAPPPRR323 pKa = 11.84TDD325 pKa = 4.28KK326 pKa = 11.16RR327 pKa = 11.84IFDD330 pKa = 4.9CILNFAYY337 pKa = 9.7KK338 pKa = 10.19FVDD341 pKa = 4.0GSVTKK346 pKa = 10.36TSLIKK351 pKa = 10.87AFTKK355 pKa = 10.51FADD358 pKa = 3.46THH360 pKa = 6.22RR361 pKa = 11.84FTIKK365 pKa = 10.44DD366 pKa = 4.09HH367 pKa = 6.83DD368 pKa = 3.89WSSYY372 pKa = 10.36GYY374 pKa = 10.4EE375 pKa = 4.24CLFCTALMNGDD386 pKa = 3.74FPKK389 pKa = 10.69VHH391 pKa = 6.6TKK393 pKa = 10.14HH394 pKa = 7.11DD395 pKa = 4.25YY396 pKa = 8.43VTSILKK402 pKa = 10.63AGGILIEE409 pKa = 4.56ILDD412 pKa = 3.74NRR414 pKa = 11.84CAYY417 pKa = 9.3RR418 pKa = 11.84KK419 pKa = 9.9KK420 pKa = 10.45YY421 pKa = 9.44PRR423 pKa = 11.84PKK425 pKa = 10.35KK426 pKa = 9.78RR427 pKa = 11.84FHH429 pKa = 7.0SNRR432 pKa = 11.84LMPGAPVPLYY442 pKa = 9.3PLMPVVHH449 pKa = 6.52TEE451 pKa = 5.22DD452 pKa = 3.03KK453 pKa = 10.34TEE455 pKa = 3.72WVTFLTGEE463 pKa = 4.65KK464 pKa = 9.73MIFHH468 pKa = 7.11NKK470 pKa = 8.0TSQCYY475 pKa = 9.37FEE477 pKa = 6.34IIGLNKK483 pKa = 10.18SPSFEE488 pKa = 4.22ITCSPQVSPDD498 pKa = 3.29CLTRR502 pKa = 11.84CYY504 pKa = 10.87KK505 pKa = 10.48FVQHH509 pKa = 6.32WLGYY513 pKa = 9.53PPGNSGNKK521 pKa = 9.15YY522 pKa = 8.48LWSRR526 pKa = 11.84QFDD529 pKa = 4.09PRR531 pKa = 11.84CCWPFDD537 pKa = 3.62SDD539 pKa = 5.39DD540 pKa = 4.8PYY542 pKa = 10.83TSKK545 pKa = 11.2PKK547 pKa = 8.73MLFVAVHH554 pKa = 6.22NIEE557 pKa = 4.05RR558 pKa = 11.84SQIEE562 pKa = 4.11IQIQPSGKK570 pKa = 9.16VLKK573 pKa = 9.57CLSRR577 pKa = 11.84ITVDD581 pKa = 3.25TKK583 pKa = 10.19VKK585 pKa = 9.85PRR587 pKa = 11.84ARR589 pKa = 11.84SLRR592 pKa = 11.84MQVLDD597 pKa = 4.02FNKK600 pKa = 10.47SGVIGQLVGSGQKK613 pKa = 8.5MQILPMPPPKK623 pKa = 9.8PWPSGFAFEE632 pKa = 4.72GAAGLDD638 pKa = 3.73PNVTFHH644 pKa = 6.91TKK646 pKa = 8.94QQVFKK651 pKa = 10.74QLVSFLLNHH660 pKa = 6.43SMPPDD665 pKa = 3.51VLLSTVLPEE674 pKa = 3.95WKK676 pKa = 8.64STFGRR681 pKa = 11.84LLGGLEE687 pKa = 3.97TLAVRR692 pKa = 11.84SHH694 pKa = 5.98RR695 pKa = 11.84HH696 pKa = 3.56GHH698 pKa = 5.65SYY700 pKa = 10.39NIEE703 pKa = 4.08YY704 pKa = 9.36MSKK707 pKa = 9.09TKK709 pKa = 10.7NISNFLRR716 pKa = 11.84TLALSVPRR724 pKa = 11.84EE725 pKa = 4.09DD726 pKa = 3.32NVTIVKK732 pKa = 9.44VNPCDD737 pKa = 3.5MVLSITDD744 pKa = 5.02DD745 pKa = 3.06ILKK748 pKa = 8.74YY749 pKa = 7.08TTSCPPMEE757 pKa = 4.16VVEE760 pKa = 4.73FVSVTKK766 pKa = 10.34EE767 pKa = 3.56PWALSSISLGPGQCNEE783 pKa = 3.66GMFLILARR791 pKa = 11.84TRR793 pKa = 11.84HH794 pKa = 5.7RR795 pKa = 11.84SVSQLEE801 pKa = 4.1GMGYY805 pKa = 9.98KK806 pKa = 9.68VEE808 pKa = 4.22KK809 pKa = 10.48QLTADD814 pKa = 3.75EE815 pKa = 4.68YY816 pKa = 11.29EE817 pKa = 4.09NLPFYY822 pKa = 9.78RR823 pKa = 11.84TLEE826 pKa = 3.85ILGKK830 pKa = 8.67FTSLGFLPLEE840 pKa = 4.37GMGTSILGIVVKK852 pKa = 10.63EE853 pKa = 4.28DD854 pKa = 3.27VACEE858 pKa = 3.85YY859 pKa = 10.36WVVSLKK865 pKa = 10.83DD866 pKa = 3.47VSTKK870 pKa = 10.22PFQIKK875 pKa = 9.34ARR877 pKa = 11.84KK878 pKa = 8.51GKK880 pKa = 9.99RR881 pKa = 11.84INIGTTTGADD891 pKa = 4.45LINEE895 pKa = 4.77AIEE898 pKa = 4.68KK899 pKa = 9.88IKK901 pKa = 10.55QRR903 pKa = 11.84NLEE906 pKa = 4.29ANKK909 pKa = 9.07STAEE913 pKa = 4.02DD914 pKa = 4.3LYY916 pKa = 11.95KK917 pKa = 10.13MVNDD921 pKa = 4.43LDD923 pKa = 5.01DD924 pKa = 3.95WDD926 pKa = 4.11TDD928 pKa = 4.31SIAGEE933 pKa = 4.11LKK935 pKa = 10.95DD936 pKa = 6.05NDD938 pKa = 4.18MAWLNDD944 pKa = 3.63RR945 pKa = 11.84VSMSDD950 pKa = 2.82RR951 pKa = 11.84ASPTRR956 pKa = 11.84TLSRR960 pKa = 11.84PGTPTSVEE968 pKa = 4.05TATASALSSWDD979 pKa = 3.3FKK981 pKa = 10.98RR982 pKa = 11.84VNRR985 pKa = 11.84RR986 pKa = 11.84LSFF989 pKa = 3.83
MM1 pKa = 6.91NHH3 pKa = 6.22RR4 pKa = 11.84MNSRR8 pKa = 11.84KK9 pKa = 9.31QKK11 pKa = 10.43HH12 pKa = 5.5GDD14 pKa = 3.02MMNYY18 pKa = 10.41NMTNIYY24 pKa = 8.03NTNAMMRR31 pKa = 11.84IRR33 pKa = 11.84KK34 pKa = 9.19RR35 pKa = 11.84KK36 pKa = 9.63INDD39 pKa = 3.53AEE41 pKa = 4.1DD42 pKa = 3.15MRR44 pKa = 11.84AKK46 pKa = 10.36RR47 pKa = 11.84KK48 pKa = 9.9CLEE51 pKa = 4.24DD52 pKa = 3.37LRR54 pKa = 11.84KK55 pKa = 9.95LRR57 pKa = 11.84TEE59 pKa = 3.63THH61 pKa = 4.78EE62 pKa = 3.92TGFRR66 pKa = 11.84IEE68 pKa = 4.06TLRR71 pKa = 11.84AAEE74 pKa = 4.83LLMEE78 pKa = 5.03DD79 pKa = 4.62AGFSDD84 pKa = 4.02STVTRR89 pKa = 11.84ANSKK93 pKa = 9.8LLRR96 pKa = 11.84NLGHH100 pKa = 7.13KK101 pKa = 9.93IKK103 pKa = 10.44PPNIQLHH110 pKa = 6.05NISMNRR116 pKa = 11.84FSSIMVKK123 pKa = 9.53TSEE126 pKa = 4.39YY127 pKa = 10.09EE128 pKa = 4.05LNSLGIEE135 pKa = 4.59PIHH138 pKa = 6.19AAGRR142 pKa = 11.84TYY144 pKa = 10.79IKK146 pKa = 10.67FQDD149 pKa = 4.02LSKK152 pKa = 10.41LCCSRR157 pKa = 11.84TTDD160 pKa = 3.67FDD162 pKa = 3.76SALAEE167 pKa = 4.3FFNLMSQRR175 pKa = 11.84TQRR178 pKa = 11.84SYY180 pKa = 11.8NLWKK184 pKa = 10.82SLVDD188 pKa = 3.26EE189 pKa = 4.5AQILEE194 pKa = 4.38VPRR197 pKa = 11.84VPTHH201 pKa = 5.25QLQEE205 pKa = 3.78VVMRR209 pKa = 11.84DD210 pKa = 3.03KK211 pKa = 11.21VLPTTSSPLEE221 pKa = 3.82VLSYY225 pKa = 9.9CFPAVYY231 pKa = 9.93PNVVRR236 pKa = 11.84LSPLAMVEE244 pKa = 3.66NDD246 pKa = 3.08AKK248 pKa = 10.74FVEE251 pKa = 4.44NVIRR255 pKa = 11.84MVKK258 pKa = 9.94SLHH261 pKa = 6.21FKK263 pKa = 9.61MLNQIDD269 pKa = 4.45GEE271 pKa = 4.65VLGHH275 pKa = 6.83HH276 pKa = 6.55LMKK279 pKa = 11.08LMTEE283 pKa = 4.35DD284 pKa = 3.46KK285 pKa = 10.94GLRR288 pKa = 11.84NCSRR292 pKa = 11.84ATPEE296 pKa = 3.79GFEE299 pKa = 4.06MFSCFFNNTYY309 pKa = 9.72EE310 pKa = 4.16SHH312 pKa = 6.69SISINMAPPPRR323 pKa = 11.84TDD325 pKa = 4.28KK326 pKa = 11.16RR327 pKa = 11.84IFDD330 pKa = 4.9CILNFAYY337 pKa = 9.7KK338 pKa = 10.19FVDD341 pKa = 4.0GSVTKK346 pKa = 10.36TSLIKK351 pKa = 10.87AFTKK355 pKa = 10.51FADD358 pKa = 3.46THH360 pKa = 6.22RR361 pKa = 11.84FTIKK365 pKa = 10.44DD366 pKa = 4.09HH367 pKa = 6.83DD368 pKa = 3.89WSSYY372 pKa = 10.36GYY374 pKa = 10.4EE375 pKa = 4.24CLFCTALMNGDD386 pKa = 3.74FPKK389 pKa = 10.69VHH391 pKa = 6.6TKK393 pKa = 10.14HH394 pKa = 7.11DD395 pKa = 4.25YY396 pKa = 8.43VTSILKK402 pKa = 10.63AGGILIEE409 pKa = 4.56ILDD412 pKa = 3.74NRR414 pKa = 11.84CAYY417 pKa = 9.3RR418 pKa = 11.84KK419 pKa = 9.9KK420 pKa = 10.45YY421 pKa = 9.44PRR423 pKa = 11.84PKK425 pKa = 10.35KK426 pKa = 9.78RR427 pKa = 11.84FHH429 pKa = 7.0SNRR432 pKa = 11.84LMPGAPVPLYY442 pKa = 9.3PLMPVVHH449 pKa = 6.52TEE451 pKa = 5.22DD452 pKa = 3.03KK453 pKa = 10.34TEE455 pKa = 3.72WVTFLTGEE463 pKa = 4.65KK464 pKa = 9.73MIFHH468 pKa = 7.11NKK470 pKa = 8.0TSQCYY475 pKa = 9.37FEE477 pKa = 6.34IIGLNKK483 pKa = 10.18SPSFEE488 pKa = 4.22ITCSPQVSPDD498 pKa = 3.29CLTRR502 pKa = 11.84CYY504 pKa = 10.87KK505 pKa = 10.48FVQHH509 pKa = 6.32WLGYY513 pKa = 9.53PPGNSGNKK521 pKa = 9.15YY522 pKa = 8.48LWSRR526 pKa = 11.84QFDD529 pKa = 4.09PRR531 pKa = 11.84CCWPFDD537 pKa = 3.62SDD539 pKa = 5.39DD540 pKa = 4.8PYY542 pKa = 10.83TSKK545 pKa = 11.2PKK547 pKa = 8.73MLFVAVHH554 pKa = 6.22NIEE557 pKa = 4.05RR558 pKa = 11.84SQIEE562 pKa = 4.11IQIQPSGKK570 pKa = 9.16VLKK573 pKa = 9.57CLSRR577 pKa = 11.84ITVDD581 pKa = 3.25TKK583 pKa = 10.19VKK585 pKa = 9.85PRR587 pKa = 11.84ARR589 pKa = 11.84SLRR592 pKa = 11.84MQVLDD597 pKa = 4.02FNKK600 pKa = 10.47SGVIGQLVGSGQKK613 pKa = 8.5MQILPMPPPKK623 pKa = 9.8PWPSGFAFEE632 pKa = 4.72GAAGLDD638 pKa = 3.73PNVTFHH644 pKa = 6.91TKK646 pKa = 8.94QQVFKK651 pKa = 10.74QLVSFLLNHH660 pKa = 6.43SMPPDD665 pKa = 3.51VLLSTVLPEE674 pKa = 3.95WKK676 pKa = 8.64STFGRR681 pKa = 11.84LLGGLEE687 pKa = 3.97TLAVRR692 pKa = 11.84SHH694 pKa = 5.98RR695 pKa = 11.84HH696 pKa = 3.56GHH698 pKa = 5.65SYY700 pKa = 10.39NIEE703 pKa = 4.08YY704 pKa = 9.36MSKK707 pKa = 9.09TKK709 pKa = 10.7NISNFLRR716 pKa = 11.84TLALSVPRR724 pKa = 11.84EE725 pKa = 4.09DD726 pKa = 3.32NVTIVKK732 pKa = 9.44VNPCDD737 pKa = 3.5MVLSITDD744 pKa = 5.02DD745 pKa = 3.06ILKK748 pKa = 8.74YY749 pKa = 7.08TTSCPPMEE757 pKa = 4.16VVEE760 pKa = 4.73FVSVTKK766 pKa = 10.34EE767 pKa = 3.56PWALSSISLGPGQCNEE783 pKa = 3.66GMFLILARR791 pKa = 11.84TRR793 pKa = 11.84HH794 pKa = 5.7RR795 pKa = 11.84SVSQLEE801 pKa = 4.1GMGYY805 pKa = 9.98KK806 pKa = 9.68VEE808 pKa = 4.22KK809 pKa = 10.48QLTADD814 pKa = 3.75EE815 pKa = 4.68YY816 pKa = 11.29EE817 pKa = 4.09NLPFYY822 pKa = 9.78RR823 pKa = 11.84TLEE826 pKa = 3.85ILGKK830 pKa = 8.67FTSLGFLPLEE840 pKa = 4.37GMGTSILGIVVKK852 pKa = 10.63EE853 pKa = 4.28DD854 pKa = 3.27VACEE858 pKa = 3.85YY859 pKa = 10.36WVVSLKK865 pKa = 10.83DD866 pKa = 3.47VSTKK870 pKa = 10.22PFQIKK875 pKa = 9.34ARR877 pKa = 11.84KK878 pKa = 8.51GKK880 pKa = 9.99RR881 pKa = 11.84INIGTTTGADD891 pKa = 4.45LINEE895 pKa = 4.77AIEE898 pKa = 4.68KK899 pKa = 9.88IKK901 pKa = 10.55QRR903 pKa = 11.84NLEE906 pKa = 4.29ANKK909 pKa = 9.07STAEE913 pKa = 4.02DD914 pKa = 4.3LYY916 pKa = 11.95KK917 pKa = 10.13MVNDD921 pKa = 4.43LDD923 pKa = 5.01DD924 pKa = 3.95WDD926 pKa = 4.11TDD928 pKa = 4.31SIAGEE933 pKa = 4.11LKK935 pKa = 10.95DD936 pKa = 6.05NDD938 pKa = 4.18MAWLNDD944 pKa = 3.63RR945 pKa = 11.84VSMSDD950 pKa = 2.82RR951 pKa = 11.84ASPTRR956 pKa = 11.84TLSRR960 pKa = 11.84PGTPTSVEE968 pKa = 4.05TATASALSSWDD979 pKa = 3.3FKK981 pKa = 10.98RR982 pKa = 11.84VNRR985 pKa = 11.84RR986 pKa = 11.84LSFF989 pKa = 3.83
Molecular weight: 112.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4252 |
244 |
1292 |
607.4 |
68.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.691 ± 0.438 | 2.117 ± 0.252 |
4.915 ± 0.19 | 5.738 ± 0.575 |
4.586 ± 0.289 | 5.456 ± 0.642 |
2.352 ± 0.437 | 5.691 ± 0.189 |
5.88 ± 0.504 | 9.737 ± 0.691 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.658 ± 0.367 | 4.516 ± 0.403 |
4.986 ± 0.257 | 3.504 ± 0.384 |
5.127 ± 0.407 | 8.984 ± 0.678 |
6.985 ± 0.447 | 6.115 ± 0.481 |
1.599 ± 0.192 | 3.293 ± 0.37 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
