
Sinocyclocheilus grahami (Dianchi golden-line fish) (Barbus grahami)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai;
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
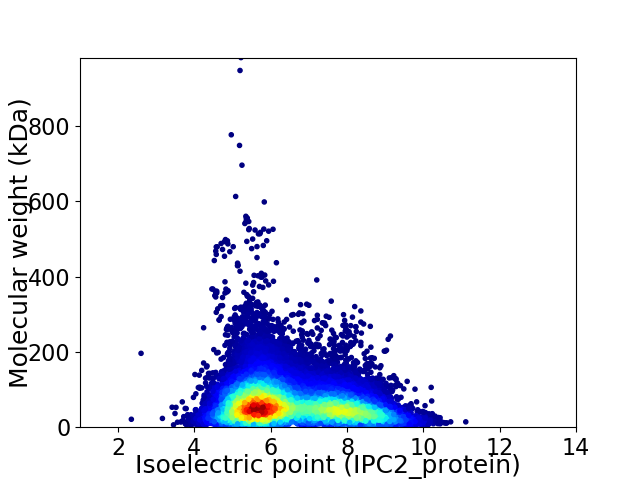
Virtual 2D-PAGE plot for 102128 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A672RYX2|A0A672RYX2_SINGR Isoform of A0A672RXC5 Myotubularin-related protein 5-like OS=Sinocyclocheilus grahami OX=75366 GN=LOC107581285 PE=3 SV=1
KKK2 pKa = 10.5HH3 pKa = 6.07ISEEE7 pKa = 4.41KKK9 pKa = 7.93TGDDD13 pKa = 3.53SFCPEEE19 pKa = 5.15GPTDDD24 pKa = 4.03KK25 pKa = 10.76CTSGQSEEE33 pKa = 4.19LGDDD37 pKa = 3.63SSSGQLEEE45 pKa = 4.07TAEEE49 pKa = 4.45FTTDDD54 pKa = 2.94SGLTGEEE61 pKa = 4.35SSSGQLGPTAEEE73 pKa = 4.88FTTDDD78 pKa = 2.94SGLTGEEE85 pKa = 4.35SSSGQLGPTAEEE97 pKa = 4.88FTTDDD102 pKa = 2.94SGLTGEEE109 pKa = 4.35SSSGQLGPTAEEE121 pKa = 5.2FTTDDD126 pKa = 3.17SGLTGEEE133 pKa = 4.35SSSGQLGPTAEEE145 pKa = 5.2FTTDDD150 pKa = 3.17SGLTGEEE157 pKa = 4.35SSSGQLGPTAEEE169 pKa = 5.2FTTDDD174 pKa = 3.17SGLTGEEE181 pKa = 4.35SSSGQLGPTAEEE193 pKa = 5.2FTTDDD198 pKa = 3.17SGLTGEEE205 pKa = 4.35SSSGQLGPTAEEE217 pKa = 5.2FTTDDD222 pKa = 3.17SGLTGEEE229 pKa = 4.35SSSGQLGPTAEEE241 pKa = 5.2FTTDDD246 pKa = 3.17SGLTGEEE253 pKa = 4.35SSSGQLGPTAEEE265 pKa = 5.2FTTDDD270 pKa = 3.17SGLTGEEE277 pKa = 4.35SSSGQLGPTAEEE289 pKa = 5.2FTTDDD294 pKa = 3.17SGLTGEEE301 pKa = 4.35SSSGQLGPTAEEE313 pKa = 5.2FTTDDD318 pKa = 3.17SGLTGEEE325 pKa = 4.35SSSGQLGPTAEEE337 pKa = 5.2FTTDDD342 pKa = 3.17SGLTGEEE349 pKa = 4.35SSSGQLGPTAEEE361 pKa = 5.2FTTDDD366 pKa = 3.17SGLTGEEE373 pKa = 4.35SSSGQLGPTAEEE385 pKa = 5.2FTTDDD390 pKa = 3.17SGLTGEEE397 pKa = 4.35SSSGQLGPTAEEE409 pKa = 5.2FTTDDD414 pKa = 3.17SGLTGEEE421 pKa = 4.35SSSGQLGPTAEEE433 pKa = 5.2FTTDDD438 pKa = 3.17SGLTGEEE445 pKa = 4.35SSSGQLGPTAEEE457 pKa = 5.2FTTDDD462 pKa = 3.17SGLTGEEE469 pKa = 4.35SSSGQLGPTAEEE481 pKa = 5.2FTTDDD486 pKa = 3.17SGLTGEEE493 pKa = 4.35SSSGQLGPTAEEE505 pKa = 5.2FTTDDD510 pKa = 3.17SGLTGEEE517 pKa = 4.35SSSGQLGPTAEEE529 pKa = 5.2FTTDDD534 pKa = 3.17SGLTGEEE541 pKa = 4.35SSSGQLGPTAEEE553 pKa = 5.2FTTDDD558 pKa = 3.17SGLTGEEE565 pKa = 4.35SSSGQLGPTSMICKKK580 pKa = 9.4SSPSRR585 pKa = 11.84RR586 pKa = 11.84YYY588 pKa = 9.47VGAHHH593 pKa = 5.98DDD595 pKa = 3.86IKKK598 pKa = 10.46HH599 pKa = 6.15GAIRR603 pKa = 11.84YYY605 pKa = 7.26GICQTLSMHHH615 pKa = 7.01LPFKKK620 pKa = 10.46LRR622 pKa = 11.84LMFLKKK628 pKa = 10.51DD629 pKa = 3.06SYYY632 pKa = 10.73YYY634 pKa = 9.99DDD636 pKa = 3.76IYYY639 pKa = 10.3IKKK642 pKa = 10.4TVQINIVKKK651 pKa = 7.77YY652 pKa = 9.67YY653 pKa = 10.74YYY655 pKa = 10.3LC
KKK2 pKa = 10.5HH3 pKa = 6.07ISEEE7 pKa = 4.41KKK9 pKa = 7.93TGDDD13 pKa = 3.53SFCPEEE19 pKa = 5.15GPTDDD24 pKa = 4.03KK25 pKa = 10.76CTSGQSEEE33 pKa = 4.19LGDDD37 pKa = 3.63SSSGQLEEE45 pKa = 4.07TAEEE49 pKa = 4.45FTTDDD54 pKa = 2.94SGLTGEEE61 pKa = 4.35SSSGQLGPTAEEE73 pKa = 4.88FTTDDD78 pKa = 2.94SGLTGEEE85 pKa = 4.35SSSGQLGPTAEEE97 pKa = 4.88FTTDDD102 pKa = 2.94SGLTGEEE109 pKa = 4.35SSSGQLGPTAEEE121 pKa = 5.2FTTDDD126 pKa = 3.17SGLTGEEE133 pKa = 4.35SSSGQLGPTAEEE145 pKa = 5.2FTTDDD150 pKa = 3.17SGLTGEEE157 pKa = 4.35SSSGQLGPTAEEE169 pKa = 5.2FTTDDD174 pKa = 3.17SGLTGEEE181 pKa = 4.35SSSGQLGPTAEEE193 pKa = 5.2FTTDDD198 pKa = 3.17SGLTGEEE205 pKa = 4.35SSSGQLGPTAEEE217 pKa = 5.2FTTDDD222 pKa = 3.17SGLTGEEE229 pKa = 4.35SSSGQLGPTAEEE241 pKa = 5.2FTTDDD246 pKa = 3.17SGLTGEEE253 pKa = 4.35SSSGQLGPTAEEE265 pKa = 5.2FTTDDD270 pKa = 3.17SGLTGEEE277 pKa = 4.35SSSGQLGPTAEEE289 pKa = 5.2FTTDDD294 pKa = 3.17SGLTGEEE301 pKa = 4.35SSSGQLGPTAEEE313 pKa = 5.2FTTDDD318 pKa = 3.17SGLTGEEE325 pKa = 4.35SSSGQLGPTAEEE337 pKa = 5.2FTTDDD342 pKa = 3.17SGLTGEEE349 pKa = 4.35SSSGQLGPTAEEE361 pKa = 5.2FTTDDD366 pKa = 3.17SGLTGEEE373 pKa = 4.35SSSGQLGPTAEEE385 pKa = 5.2FTTDDD390 pKa = 3.17SGLTGEEE397 pKa = 4.35SSSGQLGPTAEEE409 pKa = 5.2FTTDDD414 pKa = 3.17SGLTGEEE421 pKa = 4.35SSSGQLGPTAEEE433 pKa = 5.2FTTDDD438 pKa = 3.17SGLTGEEE445 pKa = 4.35SSSGQLGPTAEEE457 pKa = 5.2FTTDDD462 pKa = 3.17SGLTGEEE469 pKa = 4.35SSSGQLGPTAEEE481 pKa = 5.2FTTDDD486 pKa = 3.17SGLTGEEE493 pKa = 4.35SSSGQLGPTAEEE505 pKa = 5.2FTTDDD510 pKa = 3.17SGLTGEEE517 pKa = 4.35SSSGQLGPTAEEE529 pKa = 5.2FTTDDD534 pKa = 3.17SGLTGEEE541 pKa = 4.35SSSGQLGPTAEEE553 pKa = 5.2FTTDDD558 pKa = 3.17SGLTGEEE565 pKa = 4.35SSSGQLGPTSMICKKK580 pKa = 9.4SSPSRR585 pKa = 11.84RR586 pKa = 11.84YYY588 pKa = 9.47VGAHHH593 pKa = 5.98DDD595 pKa = 3.86IKKK598 pKa = 10.46HH599 pKa = 6.15GAIRR603 pKa = 11.84YYY605 pKa = 7.26GICQTLSMHHH615 pKa = 7.01LPFKKK620 pKa = 10.46LRR622 pKa = 11.84LMFLKKK628 pKa = 10.51DD629 pKa = 3.06SYYY632 pKa = 10.73YYY634 pKa = 9.99DDD636 pKa = 3.76IYYY639 pKa = 10.3IKKK642 pKa = 10.4TVQINIVKKK651 pKa = 7.77YY652 pKa = 9.67YY653 pKa = 10.74YYY655 pKa = 10.3LC
Molecular weight: 67.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A672MC37|A0A672MC37_SINGR A kinase (PRKA) anchor protein 7 OS=Sinocyclocheilus grahami OX=75366 PE=4 SV=1
MM1 pKa = 7.62RR2 pKa = 11.84SAAAMDD8 pKa = 3.55VGVTVLLHH16 pKa = 5.55IQVFVIFSFGQLLIVGGGGGRR37 pKa = 11.84LWPGPGQRR45 pKa = 11.84LWPGPGQRR53 pKa = 11.84LWPGPGQRR61 pKa = 11.84LWAGPGQRR69 pKa = 11.84LWSRR73 pKa = 11.84PGKK76 pKa = 10.04RR77 pKa = 11.84LWPGKK82 pKa = 9.54RR83 pKa = 11.84LWSRR87 pKa = 11.84PGKK90 pKa = 10.04RR91 pKa = 11.84LWPGKK96 pKa = 9.62RR97 pKa = 11.84LWPGKK102 pKa = 9.62RR103 pKa = 11.84LWPGKK108 pKa = 9.62RR109 pKa = 11.84LWPGKK114 pKa = 9.62RR115 pKa = 11.84LWPGKK120 pKa = 9.62RR121 pKa = 11.84LWPGKK126 pKa = 9.62RR127 pKa = 11.84LWPGKK132 pKa = 9.62RR133 pKa = 11.84LWPGKK138 pKa = 9.54RR139 pKa = 11.84LWSRR143 pKa = 11.84PGKK146 pKa = 10.04RR147 pKa = 11.84LWPGKK152 pKa = 9.54RR153 pKa = 11.84LWSRR157 pKa = 11.84PGKK160 pKa = 10.04RR161 pKa = 11.84LWPGKK166 pKa = 9.54RR167 pKa = 11.84LWSRR171 pKa = 11.84PGKK174 pKa = 10.04RR175 pKa = 11.84LWPGKK180 pKa = 10.27RR181 pKa = 11.84PGKK184 pKa = 10.32RR185 pKa = 11.84PGKK188 pKa = 9.84RR189 pKa = 11.84LWPGKK194 pKa = 8.31WLWPGKK200 pKa = 7.46WARR203 pKa = 11.84KK204 pKa = 8.0WSMKK208 pKa = 10.39
MM1 pKa = 7.62RR2 pKa = 11.84SAAAMDD8 pKa = 3.55VGVTVLLHH16 pKa = 5.55IQVFVIFSFGQLLIVGGGGGRR37 pKa = 11.84LWPGPGQRR45 pKa = 11.84LWPGPGQRR53 pKa = 11.84LWPGPGQRR61 pKa = 11.84LWAGPGQRR69 pKa = 11.84LWSRR73 pKa = 11.84PGKK76 pKa = 10.04RR77 pKa = 11.84LWPGKK82 pKa = 9.54RR83 pKa = 11.84LWSRR87 pKa = 11.84PGKK90 pKa = 10.04RR91 pKa = 11.84LWPGKK96 pKa = 9.62RR97 pKa = 11.84LWPGKK102 pKa = 9.62RR103 pKa = 11.84LWPGKK108 pKa = 9.62RR109 pKa = 11.84LWPGKK114 pKa = 9.62RR115 pKa = 11.84LWPGKK120 pKa = 9.62RR121 pKa = 11.84LWPGKK126 pKa = 9.62RR127 pKa = 11.84LWPGKK132 pKa = 9.62RR133 pKa = 11.84LWPGKK138 pKa = 9.54RR139 pKa = 11.84LWSRR143 pKa = 11.84PGKK146 pKa = 10.04RR147 pKa = 11.84LWPGKK152 pKa = 9.54RR153 pKa = 11.84LWSRR157 pKa = 11.84PGKK160 pKa = 10.04RR161 pKa = 11.84LWPGKK166 pKa = 9.54RR167 pKa = 11.84LWSRR171 pKa = 11.84PGKK174 pKa = 10.04RR175 pKa = 11.84LWPGKK180 pKa = 10.27RR181 pKa = 11.84PGKK184 pKa = 10.32RR185 pKa = 11.84PGKK188 pKa = 9.84RR189 pKa = 11.84LWPGKK194 pKa = 8.31WLWPGKK200 pKa = 7.46WARR203 pKa = 11.84KK204 pKa = 8.0WSMKK208 pKa = 10.39
Molecular weight: 24.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
62724889 |
18 |
8640 |
614.2 |
68.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.261 ± 0.007 | 2.319 ± 0.007 |
5.238 ± 0.005 | 6.816 ± 0.008 |
3.903 ± 0.006 | 6.007 ± 0.008 |
2.68 ± 0.003 | 4.885 ± 0.005 |
5.835 ± 0.008 | 9.708 ± 0.01 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.479 ± 0.003 | 4.041 ± 0.005 |
5.276 ± 0.008 | 4.661 ± 0.007 |
5.427 ± 0.005 | 8.322 ± 0.008 |
5.565 ± 0.006 | 6.426 ± 0.005 |
1.174 ± 0.003 | 2.952 ± 0.004 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
