
Hubei virga-like virus 16
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.03
Get precalculated fractions of proteins
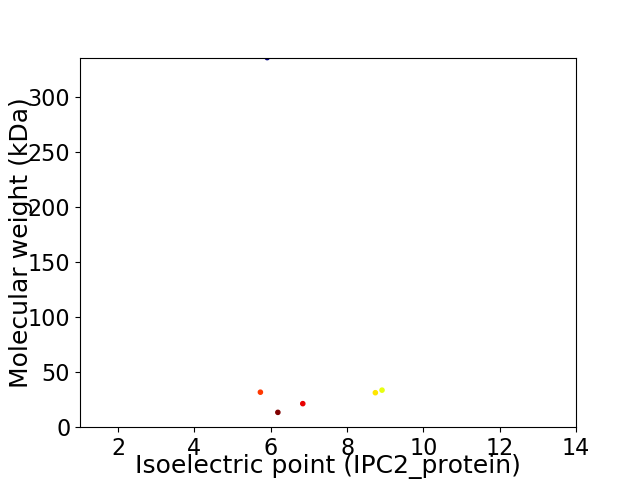
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
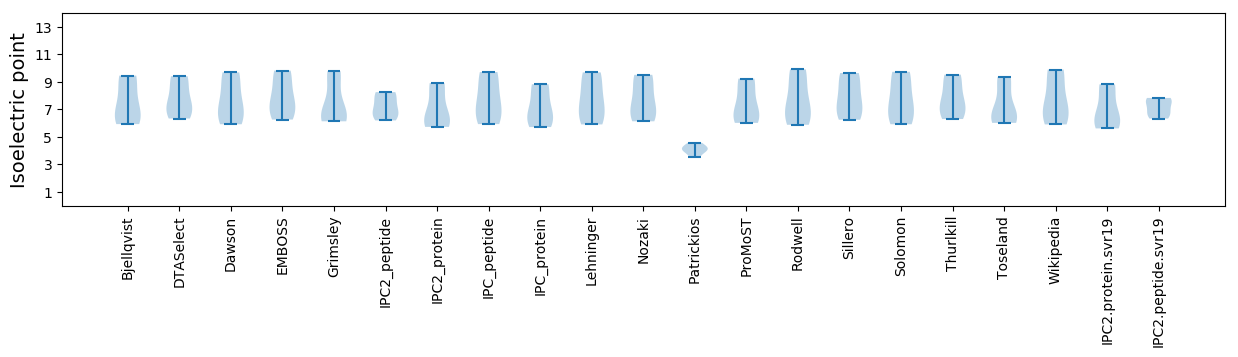
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJI0|A0A1L3KJI0_9VIRU Uncharacterized protein OS=Hubei virga-like virus 16 OX=1923331 PE=4 SV=1
MM1 pKa = 7.23VSTVLSLLNIVCLFSFGGSGFASPLVFGSNITLTDD36 pKa = 3.74FNVLLDD42 pKa = 3.87RR43 pKa = 11.84FKK45 pKa = 11.31EE46 pKa = 3.97LDD48 pKa = 3.56DD49 pKa = 4.14TVTSLAVTAVSMEE62 pKa = 4.1SMVSDD67 pKa = 4.88FIMNFQKK74 pKa = 11.07LLLQTGTHH82 pKa = 6.44FNDD85 pKa = 3.31TLIFSEE91 pKa = 4.14KK92 pKa = 9.85FYY94 pKa = 11.03NVSMQFNSLHH104 pKa = 6.31ASILTVLKK112 pKa = 9.57TLEE115 pKa = 4.13LSTTKK120 pKa = 10.38SANLSFVKK128 pKa = 10.08TFLYY132 pKa = 10.01GPKK135 pKa = 9.91GIAYY139 pKa = 8.09SIPKK143 pKa = 10.07ASYY146 pKa = 10.03QSFKK150 pKa = 10.63RR151 pKa = 11.84PSSPYY156 pKa = 8.53VTTHH160 pKa = 6.87AYY162 pKa = 8.34TSLFAGLLGDD172 pKa = 4.13IEE174 pKa = 4.5RR175 pKa = 11.84LVVNSFVYY183 pKa = 10.2VLKK186 pKa = 10.95AFLQIFIDD194 pKa = 3.89SLKK197 pKa = 10.94LIVDD201 pKa = 3.54EE202 pKa = 4.41LLRR205 pKa = 11.84VLRR208 pKa = 11.84NMLPVFEE215 pKa = 5.3RR216 pKa = 11.84LVLMLVDD223 pKa = 5.09IVQQLLKK230 pKa = 10.94LLTEE234 pKa = 4.94LFDD237 pKa = 4.82KK238 pKa = 11.25LNEE241 pKa = 4.0QYY243 pKa = 11.4FMVEE247 pKa = 3.56LVIVFGLCYY256 pKa = 10.41YY257 pKa = 7.47FTRR260 pKa = 11.84DD261 pKa = 2.73IYY263 pKa = 10.24ITIGLFLIFFSIFDD277 pKa = 3.67LTKK280 pKa = 10.55
MM1 pKa = 7.23VSTVLSLLNIVCLFSFGGSGFASPLVFGSNITLTDD36 pKa = 3.74FNVLLDD42 pKa = 3.87RR43 pKa = 11.84FKK45 pKa = 11.31EE46 pKa = 3.97LDD48 pKa = 3.56DD49 pKa = 4.14TVTSLAVTAVSMEE62 pKa = 4.1SMVSDD67 pKa = 4.88FIMNFQKK74 pKa = 11.07LLLQTGTHH82 pKa = 6.44FNDD85 pKa = 3.31TLIFSEE91 pKa = 4.14KK92 pKa = 9.85FYY94 pKa = 11.03NVSMQFNSLHH104 pKa = 6.31ASILTVLKK112 pKa = 9.57TLEE115 pKa = 4.13LSTTKK120 pKa = 10.38SANLSFVKK128 pKa = 10.08TFLYY132 pKa = 10.01GPKK135 pKa = 9.91GIAYY139 pKa = 8.09SIPKK143 pKa = 10.07ASYY146 pKa = 10.03QSFKK150 pKa = 10.63RR151 pKa = 11.84PSSPYY156 pKa = 8.53VTTHH160 pKa = 6.87AYY162 pKa = 8.34TSLFAGLLGDD172 pKa = 4.13IEE174 pKa = 4.5RR175 pKa = 11.84LVVNSFVYY183 pKa = 10.2VLKK186 pKa = 10.95AFLQIFIDD194 pKa = 3.89SLKK197 pKa = 10.94LIVDD201 pKa = 3.54EE202 pKa = 4.41LLRR205 pKa = 11.84VLRR208 pKa = 11.84NMLPVFEE215 pKa = 5.3RR216 pKa = 11.84LVLMLVDD223 pKa = 5.09IVQQLLKK230 pKa = 10.94LLTEE234 pKa = 4.94LFDD237 pKa = 4.82KK238 pKa = 11.25LNEE241 pKa = 4.0QYY243 pKa = 11.4FMVEE247 pKa = 3.56LVIVFGLCYY256 pKa = 10.41YY257 pKa = 7.47FTRR260 pKa = 11.84DD261 pKa = 2.73IYY263 pKa = 10.24ITIGLFLIFFSIFDD277 pKa = 3.67LTKK280 pKa = 10.55
Molecular weight: 31.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJH5|A0A1L3KJH5_9VIRU Helicase OS=Hubei virga-like virus 16 OX=1923331 PE=4 SV=1
MM1 pKa = 7.46NLRR4 pKa = 11.84MSTAPDD10 pKa = 3.23TVVAGKK16 pKa = 9.5PSQAKK21 pKa = 9.98VISATDD27 pKa = 3.76GAQVTRR33 pKa = 11.84AGQRR37 pKa = 11.84LEE39 pKa = 4.99DD40 pKa = 4.85PIGTAIMSCYY50 pKa = 10.42SNLVYY55 pKa = 10.8SPLIVVLYY63 pKa = 9.71VFALSCTAASIFGVEE78 pKa = 4.53GPLEE82 pKa = 3.95FAADD86 pKa = 4.94EE87 pKa = 4.09IQKK90 pKa = 9.43TYY92 pKa = 9.86NTTKK96 pKa = 10.7APAVKK101 pKa = 9.97ALAGASYY108 pKa = 10.58RR109 pKa = 11.84IVKK112 pKa = 9.64YY113 pKa = 9.98VLVYY117 pKa = 7.63QTVAITVCLVWLPYY131 pKa = 10.51AKK133 pKa = 10.28KK134 pKa = 10.08PSSKK138 pKa = 10.55NFNASVLFTVLSFLISGLGLLEE160 pKa = 5.18LFLLTQCWFLYY171 pKa = 10.33TEE173 pKa = 4.68LRR175 pKa = 11.84NPRR178 pKa = 11.84HH179 pKa = 6.07KK180 pKa = 10.86LFVAAFVGFLIIFQYY195 pKa = 10.09MADD198 pKa = 3.84PAQGQITNKK207 pKa = 9.38PKK209 pKa = 10.52RR210 pKa = 11.84IYY212 pKa = 10.12HH213 pKa = 5.37YY214 pKa = 10.77NWPTSSKK221 pKa = 9.91EE222 pKa = 3.87IEE224 pKa = 4.02FSPVKK229 pKa = 10.26FSKK232 pKa = 10.46PVVIEE237 pKa = 4.21TQTTTVFSQPVTPASTTSEE256 pKa = 4.1LANVPVKK263 pKa = 10.55HH264 pKa = 6.93SSFPYY269 pKa = 9.81SKK271 pKa = 10.69RR272 pKa = 11.84PDD274 pKa = 3.53GTSSLSRR281 pKa = 11.84SDD283 pKa = 3.21MPQIIFEE290 pKa = 4.37HH291 pKa = 5.81NRR293 pKa = 11.84RR294 pKa = 11.84KK295 pKa = 10.03NLRR298 pKa = 11.84QLPTDD303 pKa = 3.58PP304 pKa = 5.06
MM1 pKa = 7.46NLRR4 pKa = 11.84MSTAPDD10 pKa = 3.23TVVAGKK16 pKa = 9.5PSQAKK21 pKa = 9.98VISATDD27 pKa = 3.76GAQVTRR33 pKa = 11.84AGQRR37 pKa = 11.84LEE39 pKa = 4.99DD40 pKa = 4.85PIGTAIMSCYY50 pKa = 10.42SNLVYY55 pKa = 10.8SPLIVVLYY63 pKa = 9.71VFALSCTAASIFGVEE78 pKa = 4.53GPLEE82 pKa = 3.95FAADD86 pKa = 4.94EE87 pKa = 4.09IQKK90 pKa = 9.43TYY92 pKa = 9.86NTTKK96 pKa = 10.7APAVKK101 pKa = 9.97ALAGASYY108 pKa = 10.58RR109 pKa = 11.84IVKK112 pKa = 9.64YY113 pKa = 9.98VLVYY117 pKa = 7.63QTVAITVCLVWLPYY131 pKa = 10.51AKK133 pKa = 10.28KK134 pKa = 10.08PSSKK138 pKa = 10.55NFNASVLFTVLSFLISGLGLLEE160 pKa = 5.18LFLLTQCWFLYY171 pKa = 10.33TEE173 pKa = 4.68LRR175 pKa = 11.84NPRR178 pKa = 11.84HH179 pKa = 6.07KK180 pKa = 10.86LFVAAFVGFLIIFQYY195 pKa = 10.09MADD198 pKa = 3.84PAQGQITNKK207 pKa = 9.38PKK209 pKa = 10.52RR210 pKa = 11.84IYY212 pKa = 10.12HH213 pKa = 5.37YY214 pKa = 10.77NWPTSSKK221 pKa = 9.91EE222 pKa = 3.87IEE224 pKa = 4.02FSPVKK229 pKa = 10.26FSKK232 pKa = 10.46PVVIEE237 pKa = 4.21TQTTTVFSQPVTPASTTSEE256 pKa = 4.1LANVPVKK263 pKa = 10.55HH264 pKa = 6.93SSFPYY269 pKa = 9.81SKK271 pKa = 10.69RR272 pKa = 11.84PDD274 pKa = 3.53GTSSLSRR281 pKa = 11.84SDD283 pKa = 3.21MPQIIFEE290 pKa = 4.37HH291 pKa = 5.81NRR293 pKa = 11.84RR294 pKa = 11.84KK295 pKa = 10.03NLRR298 pKa = 11.84QLPTDD303 pKa = 3.58PP304 pKa = 5.06
Molecular weight: 33.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4133 |
117 |
2971 |
688.8 |
77.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.928 ± 0.734 | 2.008 ± 0.298 |
5.638 ± 1.217 | 5.444 ± 0.663 |
5.783 ± 1.971 | 5.347 ± 0.924 |
2.057 ± 0.42 | 4.936 ± 0.883 |
5.105 ± 0.382 | 10.38 ± 1.73 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.96 ± 0.296 | 3.654 ± 0.369 |
4.379 ± 0.601 | 2.976 ± 0.366 |
5.807 ± 0.815 | 7.404 ± 0.759 |
6.267 ± 0.394 | 9.654 ± 0.644 |
0.605 ± 0.151 | 4.67 ± 0.213 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
