
Staphylococcus phage vB_SscM-2
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Herelleviridae; Twortvirinae; Sciuriunavirus; unclassified Sciuriunavirus
Average proteome isoelectric point is 5.89
Get precalculated fractions of proteins
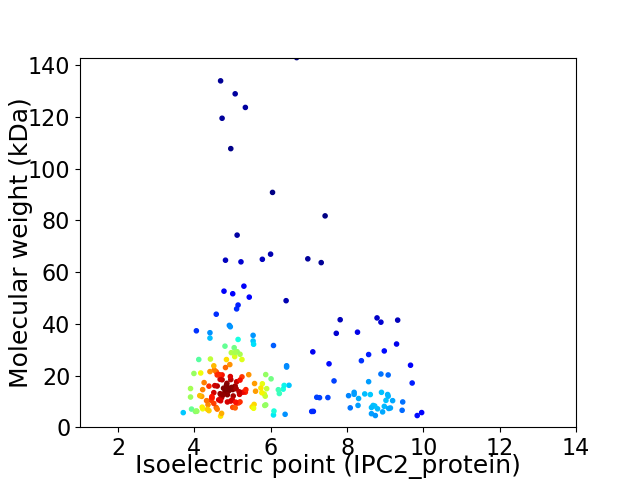
Virtual 2D-PAGE plot for 202 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1X9I9U9|A0A1X9I9U9_9CAUD Uncharacterized protein OS=Staphylococcus phage vB_SscM-2 OX=1868845 GN=vB_SscM-2_004 PE=4 SV=1
MM1 pKa = 7.51NKK3 pKa = 9.22EE4 pKa = 3.65QYY6 pKa = 10.69NYY8 pKa = 10.52LFNKK12 pKa = 7.41VTVFKK17 pKa = 11.24DD18 pKa = 3.14MEE20 pKa = 4.95DD21 pKa = 3.99YY22 pKa = 11.24NNKK25 pKa = 9.21TIDD28 pKa = 3.49GAEE31 pKa = 4.01QLDD34 pKa = 3.9EE35 pKa = 4.33ALQEE39 pKa = 4.77GFIQDD44 pKa = 3.9LTRR47 pKa = 11.84GVYY50 pKa = 10.28NGATDD55 pKa = 3.79TFTPNTLNAKK65 pKa = 9.32EE66 pKa = 4.21LVLAVQQYY74 pKa = 10.9LYY76 pKa = 9.44EE77 pKa = 4.38TDD79 pKa = 3.43WLSTLYY85 pKa = 9.2TQHH88 pKa = 6.93EE89 pKa = 4.38EE90 pKa = 4.5DD91 pKa = 5.21DD92 pKa = 4.03DD93 pKa = 5.6LEE95 pKa = 4.38TFEE98 pKa = 4.28QLTRR102 pKa = 11.84FDD104 pKa = 3.68MLEE107 pKa = 4.65DD108 pKa = 3.6YY109 pKa = 10.9LNCYY113 pKa = 10.31SVFTIGNRR121 pKa = 11.84IYY123 pKa = 11.47ADD125 pKa = 3.08ISS127 pKa = 3.42
MM1 pKa = 7.51NKK3 pKa = 9.22EE4 pKa = 3.65QYY6 pKa = 10.69NYY8 pKa = 10.52LFNKK12 pKa = 7.41VTVFKK17 pKa = 11.24DD18 pKa = 3.14MEE20 pKa = 4.95DD21 pKa = 3.99YY22 pKa = 11.24NNKK25 pKa = 9.21TIDD28 pKa = 3.49GAEE31 pKa = 4.01QLDD34 pKa = 3.9EE35 pKa = 4.33ALQEE39 pKa = 4.77GFIQDD44 pKa = 3.9LTRR47 pKa = 11.84GVYY50 pKa = 10.28NGATDD55 pKa = 3.79TFTPNTLNAKK65 pKa = 9.32EE66 pKa = 4.21LVLAVQQYY74 pKa = 10.9LYY76 pKa = 9.44EE77 pKa = 4.38TDD79 pKa = 3.43WLSTLYY85 pKa = 9.2TQHH88 pKa = 6.93EE89 pKa = 4.38EE90 pKa = 4.5DD91 pKa = 5.21DD92 pKa = 4.03DD93 pKa = 5.6LEE95 pKa = 4.38TFEE98 pKa = 4.28QLTRR102 pKa = 11.84FDD104 pKa = 3.68MLEE107 pKa = 4.65DD108 pKa = 3.6YY109 pKa = 10.9LNCYY113 pKa = 10.31SVFTIGNRR121 pKa = 11.84IYY123 pKa = 11.47ADD125 pKa = 3.08ISS127 pKa = 3.42
Molecular weight: 14.95 kDa
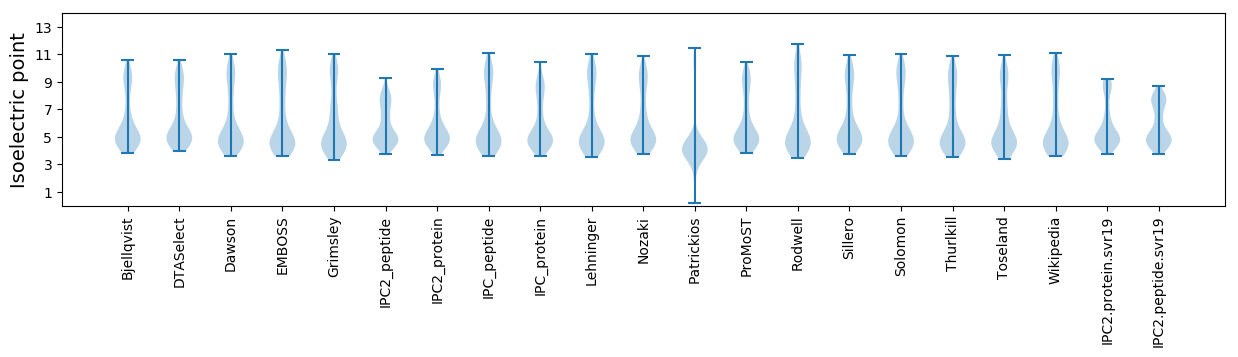
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X9IAA5|A0A1X9IAA5_9CAUD Uncharacterized protein OS=Staphylococcus phage vB_SscM-2 OX=1868845 GN=vB_SscM-2_091 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 10.39NRR4 pKa = 11.84NGRR7 pKa = 11.84NNIQHH12 pKa = 5.74QPVNFAPTNLLGGTGNNSFYY32 pKa = 10.67KK33 pKa = 10.3KK34 pKa = 10.44KK35 pKa = 10.32PGEE38 pKa = 4.37HH39 pKa = 6.77KK40 pKa = 10.76DD41 pKa = 3.36GAKK44 pKa = 9.87TIVYY48 pKa = 9.97KK49 pKa = 11.05LLFTKK54 pKa = 10.59RR55 pKa = 11.84FDD57 pKa = 3.66YY58 pKa = 11.04VSQKK62 pKa = 10.39DD63 pKa = 3.16IQMQKK68 pKa = 10.41KK69 pKa = 9.14YY70 pKa = 10.74ALNLISDD77 pKa = 3.81SLGIKK82 pKa = 10.26EE83 pKa = 4.52EE84 pKa = 3.98FLTLKK89 pKa = 10.65QKK91 pKa = 10.16GKK93 pKa = 9.86KK94 pKa = 9.28VEE96 pKa = 4.78EE97 pKa = 4.02ILHH100 pKa = 5.78TDD102 pKa = 2.76RR103 pKa = 11.84VFYY106 pKa = 8.54VHH108 pKa = 7.28RR109 pKa = 11.84GKK111 pKa = 10.87KK112 pKa = 10.35LIGKK116 pKa = 9.5CSIRR120 pKa = 11.84EE121 pKa = 3.65QRR123 pKa = 11.84TFKK126 pKa = 9.73GTHH129 pKa = 6.73LIYY132 pKa = 10.67VFKK135 pKa = 10.32TRR137 pKa = 11.84HH138 pKa = 4.77RR139 pKa = 11.84TSGKK143 pKa = 10.38KK144 pKa = 9.8KK145 pKa = 9.84GQQ147 pKa = 3.24
MM1 pKa = 7.67KK2 pKa = 10.39NRR4 pKa = 11.84NGRR7 pKa = 11.84NNIQHH12 pKa = 5.74QPVNFAPTNLLGGTGNNSFYY32 pKa = 10.67KK33 pKa = 10.3KK34 pKa = 10.44KK35 pKa = 10.32PGEE38 pKa = 4.37HH39 pKa = 6.77KK40 pKa = 10.76DD41 pKa = 3.36GAKK44 pKa = 9.87TIVYY48 pKa = 9.97KK49 pKa = 11.05LLFTKK54 pKa = 10.59RR55 pKa = 11.84FDD57 pKa = 3.66YY58 pKa = 11.04VSQKK62 pKa = 10.39DD63 pKa = 3.16IQMQKK68 pKa = 10.41KK69 pKa = 9.14YY70 pKa = 10.74ALNLISDD77 pKa = 3.81SLGIKK82 pKa = 10.26EE83 pKa = 4.52EE84 pKa = 3.98FLTLKK89 pKa = 10.65QKK91 pKa = 10.16GKK93 pKa = 9.86KK94 pKa = 9.28VEE96 pKa = 4.78EE97 pKa = 4.02ILHH100 pKa = 5.78TDD102 pKa = 2.76RR103 pKa = 11.84VFYY106 pKa = 8.54VHH108 pKa = 7.28RR109 pKa = 11.84GKK111 pKa = 10.87KK112 pKa = 10.35LIGKK116 pKa = 9.5CSIRR120 pKa = 11.84EE121 pKa = 3.65QRR123 pKa = 11.84TFKK126 pKa = 9.73GTHH129 pKa = 6.73LIYY132 pKa = 10.67VFKK135 pKa = 10.32TRR137 pKa = 11.84HH138 pKa = 4.77RR139 pKa = 11.84TSGKK143 pKa = 10.38KK144 pKa = 9.8KK145 pKa = 9.84GQQ147 pKa = 3.24
Molecular weight: 17.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
41063 |
37 |
1321 |
203.3 |
23.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.135 ± 0.177 | 0.594 ± 0.067 |
6.673 ± 0.147 | 8.808 ± 0.271 |
3.921 ± 0.107 | 5.891 ± 0.305 |
1.617 ± 0.104 | 7.411 ± 0.197 |
8.993 ± 0.255 | 8.044 ± 0.197 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.521 ± 0.106 | 6.322 ± 0.126 |
2.771 ± 0.159 | 3.331 ± 0.164 |
3.806 ± 0.11 | 6.359 ± 0.162 |
6.198 ± 0.217 | 6.592 ± 0.16 |
0.852 ± 0.057 | 5.16 ± 0.18 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
