
Myripristis murdjan (pinecone soldierfish)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha;
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
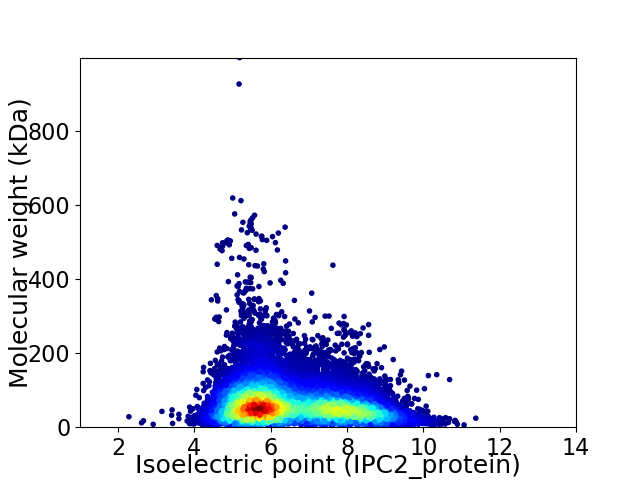
Virtual 2D-PAGE plot for 55955 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A667X8U4|A0A667X8U4_9TELE Mucolipin-2-like OS=Myripristis murdjan OX=586833 GN=LOC115358416 PE=4 SV=1
MM1 pKa = 7.74LPRR4 pKa = 11.84EE5 pKa = 4.29KK6 pKa = 10.62KK7 pKa = 10.16LLQPSEE13 pKa = 4.21PALSSVYY20 pKa = 10.3VEE22 pKa = 4.46EE23 pKa = 4.2PLYY26 pKa = 10.68PNDD29 pKa = 3.43GKK31 pKa = 9.98PWSKK35 pKa = 10.86CSLDD39 pKa = 3.77PSSKK43 pKa = 10.61SLQQSSGISSGGSSLGYY60 pKa = 10.61ANSGPDD66 pKa = 3.5IIASIQEE73 pKa = 4.43ALCKK77 pKa = 10.25ALPQLGQKK85 pKa = 10.32LPEE88 pKa = 4.86ADD90 pKa = 3.69AFLTEE95 pKa = 4.49SNKK98 pKa = 10.22NNSLLSTDD106 pKa = 4.04YY107 pKa = 10.89SPCSGRR113 pKa = 11.84DD114 pKa = 3.4DD115 pKa = 4.15DD116 pKa = 6.02LKK118 pKa = 11.44DD119 pKa = 3.66SGSSSFDD126 pKa = 2.93NRR128 pKa = 11.84TYY130 pKa = 10.96SILIPSIEE138 pKa = 4.14SQITSDD144 pKa = 3.09SSEE147 pKa = 3.99IQTQPAVICDD157 pKa = 3.57PAYY160 pKa = 10.29QPSEE164 pKa = 4.1GDD166 pKa = 3.6VMRR169 pKa = 11.84STSPDD174 pKa = 3.04QVVPVCLAAGQQDD187 pKa = 4.05AEE189 pKa = 4.57LPPAASSGLQTDD201 pKa = 5.28FSYY204 pKa = 11.04QPCNADD210 pKa = 3.41SGSSSSADD218 pKa = 3.31TSSLSSTSSDD228 pKa = 2.75INIAASCQIGAGEE241 pKa = 4.13EE242 pKa = 4.17DD243 pKa = 3.77GCEE246 pKa = 4.08DD247 pKa = 3.67SHH249 pKa = 6.73EE250 pKa = 4.43ADD252 pKa = 3.38SGAVNPAEE260 pKa = 4.2TSQDD264 pKa = 3.29VCGEE268 pKa = 3.95QAPVCDD274 pKa = 5.09ANPCYY279 pKa = 10.58GSLPAFSHH287 pKa = 6.65SSPIMDD293 pKa = 5.82DD294 pKa = 4.17DD295 pKa = 4.06YY296 pKa = 11.82QAFQSLVKK304 pKa = 10.73QPDD307 pKa = 3.11IALLEE312 pKa = 4.35EE313 pKa = 4.39SSGDD317 pKa = 3.34QWEE320 pKa = 4.24QPQLDD325 pKa = 3.84KK326 pKa = 11.31HH327 pKa = 6.18PEE329 pKa = 3.81EE330 pKa = 4.96SSTKK334 pKa = 9.7ISQSSSDD341 pKa = 3.26AVLPDD346 pKa = 4.37FSDD349 pKa = 3.25NAQAGRR355 pKa = 11.84CLPEE359 pKa = 4.15FEE361 pKa = 4.61TSLFSFLPSDD371 pKa = 3.21HH372 pKa = 6.95SEE374 pKa = 3.93AVITDD379 pKa = 3.66IEE381 pKa = 4.49SCAEE385 pKa = 3.94SGRR388 pKa = 11.84KK389 pKa = 8.66RR390 pKa = 11.84CHH392 pKa = 6.71ADD394 pKa = 3.11EE395 pKa = 5.63LDD397 pKa = 3.7LTRR400 pKa = 11.84QPLCFLFNN408 pKa = 3.96
MM1 pKa = 7.74LPRR4 pKa = 11.84EE5 pKa = 4.29KK6 pKa = 10.62KK7 pKa = 10.16LLQPSEE13 pKa = 4.21PALSSVYY20 pKa = 10.3VEE22 pKa = 4.46EE23 pKa = 4.2PLYY26 pKa = 10.68PNDD29 pKa = 3.43GKK31 pKa = 9.98PWSKK35 pKa = 10.86CSLDD39 pKa = 3.77PSSKK43 pKa = 10.61SLQQSSGISSGGSSLGYY60 pKa = 10.61ANSGPDD66 pKa = 3.5IIASIQEE73 pKa = 4.43ALCKK77 pKa = 10.25ALPQLGQKK85 pKa = 10.32LPEE88 pKa = 4.86ADD90 pKa = 3.69AFLTEE95 pKa = 4.49SNKK98 pKa = 10.22NNSLLSTDD106 pKa = 4.04YY107 pKa = 10.89SPCSGRR113 pKa = 11.84DD114 pKa = 3.4DD115 pKa = 4.15DD116 pKa = 6.02LKK118 pKa = 11.44DD119 pKa = 3.66SGSSSFDD126 pKa = 2.93NRR128 pKa = 11.84TYY130 pKa = 10.96SILIPSIEE138 pKa = 4.14SQITSDD144 pKa = 3.09SSEE147 pKa = 3.99IQTQPAVICDD157 pKa = 3.57PAYY160 pKa = 10.29QPSEE164 pKa = 4.1GDD166 pKa = 3.6VMRR169 pKa = 11.84STSPDD174 pKa = 3.04QVVPVCLAAGQQDD187 pKa = 4.05AEE189 pKa = 4.57LPPAASSGLQTDD201 pKa = 5.28FSYY204 pKa = 11.04QPCNADD210 pKa = 3.41SGSSSSADD218 pKa = 3.31TSSLSSTSSDD228 pKa = 2.75INIAASCQIGAGEE241 pKa = 4.13EE242 pKa = 4.17DD243 pKa = 3.77GCEE246 pKa = 4.08DD247 pKa = 3.67SHH249 pKa = 6.73EE250 pKa = 4.43ADD252 pKa = 3.38SGAVNPAEE260 pKa = 4.2TSQDD264 pKa = 3.29VCGEE268 pKa = 3.95QAPVCDD274 pKa = 5.09ANPCYY279 pKa = 10.58GSLPAFSHH287 pKa = 6.65SSPIMDD293 pKa = 5.82DD294 pKa = 4.17DD295 pKa = 4.06YY296 pKa = 11.82QAFQSLVKK304 pKa = 10.73QPDD307 pKa = 3.11IALLEE312 pKa = 4.35EE313 pKa = 4.39SSGDD317 pKa = 3.34QWEE320 pKa = 4.24QPQLDD325 pKa = 3.84KK326 pKa = 11.31HH327 pKa = 6.18PEE329 pKa = 3.81EE330 pKa = 4.96SSTKK334 pKa = 9.7ISQSSSDD341 pKa = 3.26AVLPDD346 pKa = 4.37FSDD349 pKa = 3.25NAQAGRR355 pKa = 11.84CLPEE359 pKa = 4.15FEE361 pKa = 4.61TSLFSFLPSDD371 pKa = 3.21HH372 pKa = 6.95SEE374 pKa = 3.93AVITDD379 pKa = 3.66IEE381 pKa = 4.49SCAEE385 pKa = 3.94SGRR388 pKa = 11.84KK389 pKa = 8.66RR390 pKa = 11.84CHH392 pKa = 6.71ADD394 pKa = 3.11EE395 pKa = 5.63LDD397 pKa = 3.7LTRR400 pKa = 11.84QPLCFLFNN408 pKa = 3.96
Molecular weight: 43.29 kDa
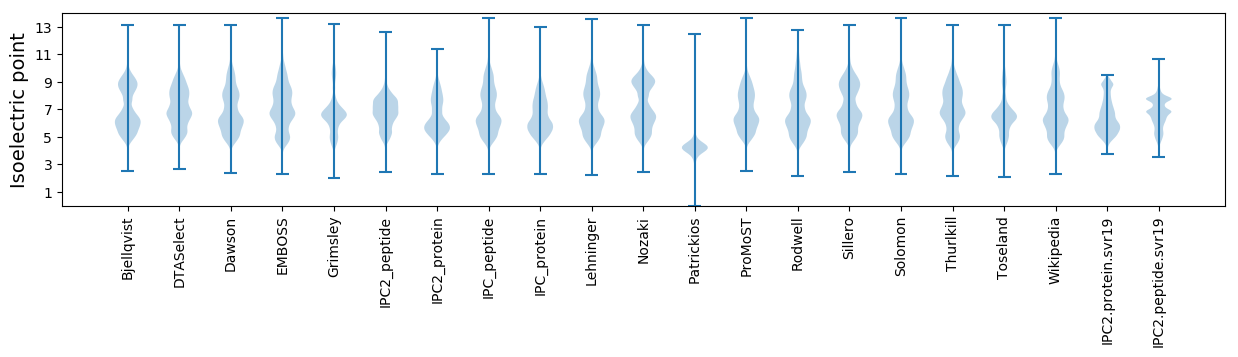
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A668AH03|A0A668AH03_9TELE Sphingosine-1-phosphate phosphatase 1-like OS=Myripristis murdjan OX=586833 GN=LOC115355965 PE=4 SV=1
SS1 pKa = 6.49SNRR4 pKa = 11.84RR5 pKa = 11.84QTSNSRR11 pKa = 11.84QNSTSRR17 pKa = 11.84QIFSNRR23 pKa = 11.84QTSTSRR29 pKa = 11.84QISTSRR35 pKa = 11.84QNSTSRR41 pKa = 11.84QISSNRR47 pKa = 11.84QTSTSRR53 pKa = 11.84QISSKK58 pKa = 10.05RR59 pKa = 11.84QTSTSRR65 pKa = 11.84QTSSKK70 pKa = 9.79RR71 pKa = 11.84QTSTSRR77 pKa = 11.84QTSSLQTSTSCRR89 pKa = 11.84QNSTSRR95 pKa = 11.84QISSNRR101 pKa = 11.84QTSTSRR107 pKa = 11.84QTSSKK112 pKa = 9.79RR113 pKa = 11.84QTSTSRR119 pKa = 11.84QTSSKK124 pKa = 9.79RR125 pKa = 11.84QTSTSRR131 pKa = 11.84QTSILQTSTSWQTSTSRR148 pKa = 11.84QTSSKK153 pKa = 9.79RR154 pKa = 11.84QTSTSRR160 pKa = 11.84QNSTSRR166 pKa = 11.84QISSNRR172 pKa = 11.84QTSTSRR178 pKa = 11.84QTSSKK183 pKa = 9.79RR184 pKa = 11.84QTSTSRR190 pKa = 11.84QTSSLQTSTSWQKK203 pKa = 11.08LLQPANLHH211 pKa = 5.92QPTNLL216 pKa = 3.41
SS1 pKa = 6.49SNRR4 pKa = 11.84RR5 pKa = 11.84QTSNSRR11 pKa = 11.84QNSTSRR17 pKa = 11.84QIFSNRR23 pKa = 11.84QTSTSRR29 pKa = 11.84QISTSRR35 pKa = 11.84QNSTSRR41 pKa = 11.84QISSNRR47 pKa = 11.84QTSTSRR53 pKa = 11.84QISSKK58 pKa = 10.05RR59 pKa = 11.84QTSTSRR65 pKa = 11.84QTSSKK70 pKa = 9.79RR71 pKa = 11.84QTSTSRR77 pKa = 11.84QTSSLQTSTSCRR89 pKa = 11.84QNSTSRR95 pKa = 11.84QISSNRR101 pKa = 11.84QTSTSRR107 pKa = 11.84QTSSKK112 pKa = 9.79RR113 pKa = 11.84QTSTSRR119 pKa = 11.84QTSSKK124 pKa = 9.79RR125 pKa = 11.84QTSTSRR131 pKa = 11.84QTSILQTSTSWQTSTSRR148 pKa = 11.84QTSSKK153 pKa = 9.79RR154 pKa = 11.84QTSTSRR160 pKa = 11.84QNSTSRR166 pKa = 11.84QISSNRR172 pKa = 11.84QTSTSRR178 pKa = 11.84QTSSKK183 pKa = 9.79RR184 pKa = 11.84QTSTSRR190 pKa = 11.84QTSSLQTSTSWQKK203 pKa = 11.08LLQPANLHH211 pKa = 5.92QPTNLL216 pKa = 3.41
Molecular weight: 24.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
34307100 |
18 |
8699 |
613.1 |
68.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.686 ± 0.008 | 2.339 ± 0.008 |
5.187 ± 0.007 | 6.758 ± 0.012 |
3.825 ± 0.006 | 6.292 ± 0.011 |
2.69 ± 0.005 | 4.578 ± 0.007 |
5.596 ± 0.011 | 9.877 ± 0.012 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.42 ± 0.005 | 3.802 ± 0.006 |
5.406 ± 0.012 | 4.678 ± 0.009 |
5.603 ± 0.007 | 8.135 ± 0.012 |
5.513 ± 0.007 | 6.512 ± 0.009 |
1.198 ± 0.003 | 2.902 ± 0.005 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
