
Turnip vein-clearing virus (TVCV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Tobamovirus
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
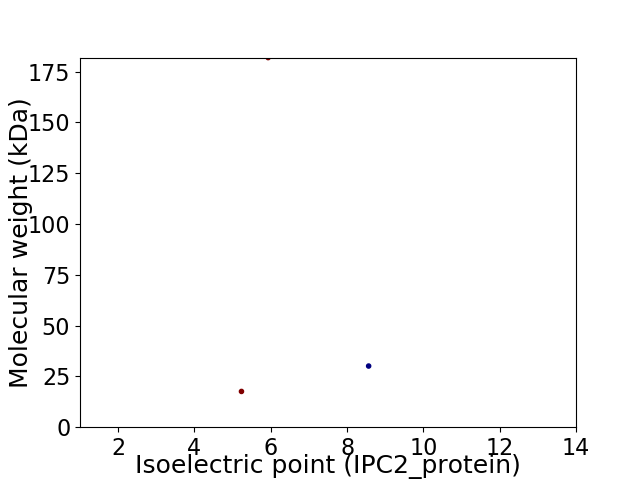
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q88922|CAPSD_TVCV Capsid protein OS=Turnip vein-clearing virus OX=29272 GN=CP PE=3 SV=3
MM1 pKa = 7.8SYY3 pKa = 11.16NITNPNQYY11 pKa = 10.54QYY13 pKa = 11.16FAAVWAEE20 pKa = 4.56PIPMLNQCMSALSQSYY36 pKa = 7.17QTQAARR42 pKa = 11.84DD43 pKa = 3.67TVRR46 pKa = 11.84QQFSNLLSAVVTPSQRR62 pKa = 11.84FPDD65 pKa = 3.64TGSRR69 pKa = 11.84VYY71 pKa = 11.19VNSAVIKK78 pKa = 9.24PLYY81 pKa = 8.18EE82 pKa = 5.16ALMKK86 pKa = 11.02SFDD89 pKa = 3.38TRR91 pKa = 11.84NRR93 pKa = 11.84IIEE96 pKa = 4.26TEE98 pKa = 4.13EE99 pKa = 3.68EE100 pKa = 4.31SRR102 pKa = 11.84PSASEE107 pKa = 3.81VANATQRR114 pKa = 11.84VDD116 pKa = 3.42DD117 pKa = 3.9ATVAIRR123 pKa = 11.84SQIQLLLSEE132 pKa = 4.98LSNGHH137 pKa = 6.27GYY139 pKa = 8.47MNRR142 pKa = 11.84AEE144 pKa = 5.04FEE146 pKa = 4.26ALLPWTTAPATT157 pKa = 3.64
MM1 pKa = 7.8SYY3 pKa = 11.16NITNPNQYY11 pKa = 10.54QYY13 pKa = 11.16FAAVWAEE20 pKa = 4.56PIPMLNQCMSALSQSYY36 pKa = 7.17QTQAARR42 pKa = 11.84DD43 pKa = 3.67TVRR46 pKa = 11.84QQFSNLLSAVVTPSQRR62 pKa = 11.84FPDD65 pKa = 3.64TGSRR69 pKa = 11.84VYY71 pKa = 11.19VNSAVIKK78 pKa = 9.24PLYY81 pKa = 8.18EE82 pKa = 5.16ALMKK86 pKa = 11.02SFDD89 pKa = 3.38TRR91 pKa = 11.84NRR93 pKa = 11.84IIEE96 pKa = 4.26TEE98 pKa = 4.13EE99 pKa = 3.68EE100 pKa = 4.31SRR102 pKa = 11.84PSASEE107 pKa = 3.81VANATQRR114 pKa = 11.84VDD116 pKa = 3.42DD117 pKa = 3.9ATVAIRR123 pKa = 11.84SQIQLLLSEE132 pKa = 4.98LSNGHH137 pKa = 6.27GYY139 pKa = 8.47MNRR142 pKa = 11.84AEE144 pKa = 5.04FEE146 pKa = 4.26ALLPWTTAPATT157 pKa = 3.64
Molecular weight: 17.6 kDa
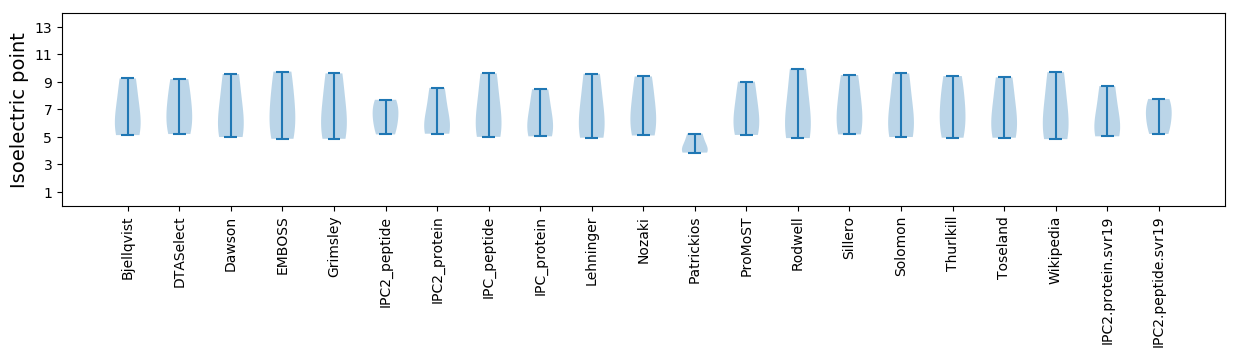
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q88922|CAPSD_TVCV Capsid protein OS=Turnip vein-clearing virus OX=29272 GN=CP PE=3 SV=3
MM1 pKa = 7.52SIVSYY6 pKa = 9.68EE7 pKa = 4.04PKK9 pKa = 10.66VSDD12 pKa = 4.59FLNLSKK18 pKa = 10.84KK19 pKa = 10.27EE20 pKa = 4.02EE21 pKa = 4.19ILPKK25 pKa = 10.55ALTRR29 pKa = 11.84LKK31 pKa = 9.72TVSISTKK38 pKa = 10.34DD39 pKa = 3.21IISVKK44 pKa = 10.06EE45 pKa = 4.02SEE47 pKa = 4.51TLCDD51 pKa = 3.78IDD53 pKa = 5.79LLINVPLDD61 pKa = 2.98KK62 pKa = 10.73YY63 pKa = 10.55RR64 pKa = 11.84YY65 pKa = 8.77VGILGAVFTGEE76 pKa = 3.81WLVPDD81 pKa = 4.61FVKK84 pKa = 10.95GGVTISVIDD93 pKa = 3.6KK94 pKa = 10.31RR95 pKa = 11.84LVNSKK100 pKa = 9.56EE101 pKa = 4.27CVIGTYY107 pKa = 10.02RR108 pKa = 11.84AAAKK112 pKa = 10.17SKK114 pKa = 9.9RR115 pKa = 11.84FQFKK119 pKa = 10.1LVPNYY124 pKa = 9.64FVSTVDD130 pKa = 3.6AKK132 pKa = 10.41RR133 pKa = 11.84KK134 pKa = 7.14PWQVHH139 pKa = 5.15VRR141 pKa = 11.84IQDD144 pKa = 3.65LKK146 pKa = 10.98IEE148 pKa = 4.82AGWQPLALEE157 pKa = 4.52VVSVAMVTNNVVMKK171 pKa = 9.81GLRR174 pKa = 11.84EE175 pKa = 3.91KK176 pKa = 10.85VVAINDD182 pKa = 3.79PDD184 pKa = 3.79VEE186 pKa = 4.25GFEE189 pKa = 4.43GVVDD193 pKa = 3.83EE194 pKa = 5.46FVDD197 pKa = 4.16SVAAFKK203 pKa = 10.95AVDD206 pKa = 3.38NFRR209 pKa = 11.84KK210 pKa = 9.79RR211 pKa = 11.84KK212 pKa = 9.58KK213 pKa = 9.95KK214 pKa = 9.75VEE216 pKa = 3.92EE217 pKa = 3.86RR218 pKa = 11.84DD219 pKa = 3.54VVSKK223 pKa = 10.67YY224 pKa = 10.41KK225 pKa = 10.53YY226 pKa = 10.04RR227 pKa = 11.84PEE229 pKa = 4.79KK230 pKa = 10.4YY231 pKa = 10.16AGPDD235 pKa = 3.12SFNLKK240 pKa = 10.02EE241 pKa = 4.15EE242 pKa = 4.36NVLQHH247 pKa = 6.13YY248 pKa = 9.65KK249 pKa = 10.35PEE251 pKa = 4.3SVPVLRR257 pKa = 11.84SGVGRR262 pKa = 11.84AHH264 pKa = 6.43TNAA267 pKa = 4.02
MM1 pKa = 7.52SIVSYY6 pKa = 9.68EE7 pKa = 4.04PKK9 pKa = 10.66VSDD12 pKa = 4.59FLNLSKK18 pKa = 10.84KK19 pKa = 10.27EE20 pKa = 4.02EE21 pKa = 4.19ILPKK25 pKa = 10.55ALTRR29 pKa = 11.84LKK31 pKa = 9.72TVSISTKK38 pKa = 10.34DD39 pKa = 3.21IISVKK44 pKa = 10.06EE45 pKa = 4.02SEE47 pKa = 4.51TLCDD51 pKa = 3.78IDD53 pKa = 5.79LLINVPLDD61 pKa = 2.98KK62 pKa = 10.73YY63 pKa = 10.55RR64 pKa = 11.84YY65 pKa = 8.77VGILGAVFTGEE76 pKa = 3.81WLVPDD81 pKa = 4.61FVKK84 pKa = 10.95GGVTISVIDD93 pKa = 3.6KK94 pKa = 10.31RR95 pKa = 11.84LVNSKK100 pKa = 9.56EE101 pKa = 4.27CVIGTYY107 pKa = 10.02RR108 pKa = 11.84AAAKK112 pKa = 10.17SKK114 pKa = 9.9RR115 pKa = 11.84FQFKK119 pKa = 10.1LVPNYY124 pKa = 9.64FVSTVDD130 pKa = 3.6AKK132 pKa = 10.41RR133 pKa = 11.84KK134 pKa = 7.14PWQVHH139 pKa = 5.15VRR141 pKa = 11.84IQDD144 pKa = 3.65LKK146 pKa = 10.98IEE148 pKa = 4.82AGWQPLALEE157 pKa = 4.52VVSVAMVTNNVVMKK171 pKa = 9.81GLRR174 pKa = 11.84EE175 pKa = 3.91KK176 pKa = 10.85VVAINDD182 pKa = 3.79PDD184 pKa = 3.79VEE186 pKa = 4.25GFEE189 pKa = 4.43GVVDD193 pKa = 3.83EE194 pKa = 5.46FVDD197 pKa = 4.16SVAAFKK203 pKa = 10.95AVDD206 pKa = 3.38NFRR209 pKa = 11.84KK210 pKa = 9.79RR211 pKa = 11.84KK212 pKa = 9.58KK213 pKa = 9.95KK214 pKa = 9.75VEE216 pKa = 3.92EE217 pKa = 3.86RR218 pKa = 11.84DD219 pKa = 3.54VVSKK223 pKa = 10.67YY224 pKa = 10.41KK225 pKa = 10.53YY226 pKa = 10.04RR227 pKa = 11.84PEE229 pKa = 4.79KK230 pKa = 10.4YY231 pKa = 10.16AGPDD235 pKa = 3.12SFNLKK240 pKa = 10.02EE241 pKa = 4.15EE242 pKa = 4.36NVLQHH247 pKa = 6.13YY248 pKa = 9.65KK249 pKa = 10.35PEE251 pKa = 4.3SVPVLRR257 pKa = 11.84SGVGRR262 pKa = 11.84AHH264 pKa = 6.43TNAA267 pKa = 4.02
Molecular weight: 30.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2025 |
157 |
1601 |
675.0 |
76.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.358 ± 0.998 | 1.975 ± 0.639 |
6.173 ± 0.807 | 6.37 ± 0.255 |
5.136 ± 0.717 | 4.346 ± 0.593 |
2.123 ± 0.606 | 5.185 ± 0.183 |
7.358 ± 1.593 | 8.938 ± 0.696 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.42 ± 0.412 | 4.395 ± 0.489 |
3.951 ± 0.563 | 3.407 ± 1.033 |
4.741 ± 0.515 | 6.765 ± 0.893 |
5.679 ± 0.668 | 8.741 ± 1.862 |
1.136 ± 0.035 | 3.802 ± 0.18 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
