
Marbled eel polyomavirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; unclassified Polyomaviridae
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
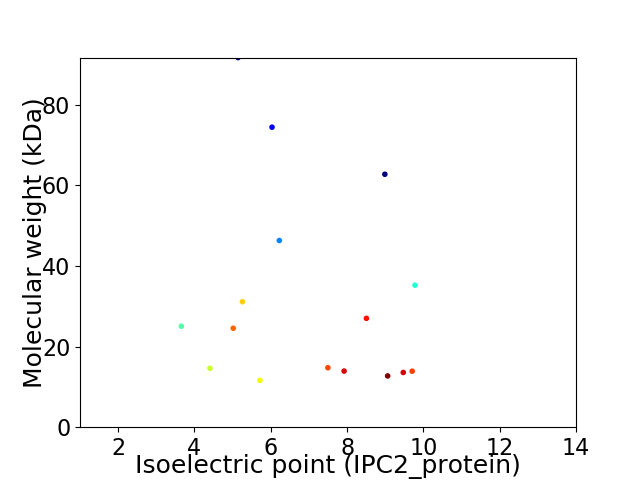
Virtual 2D-PAGE plot for 16 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
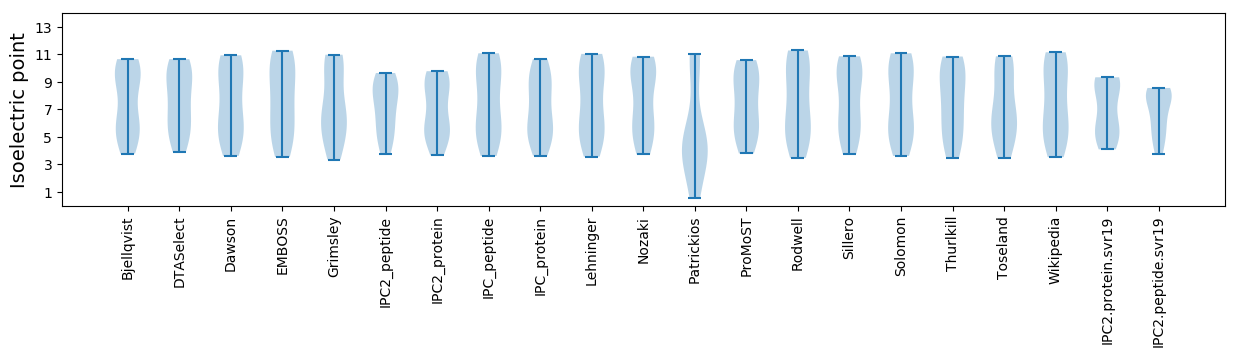
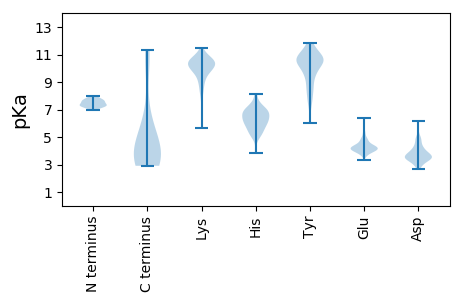
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A161IWK5|A0A161IWK5_9POLY ORF02R OS=Marbled eel polyomavirus OX=1662286 PE=4 SV=1
MM1 pKa = 7.79ASQPKK6 pKa = 9.76RR7 pKa = 11.84FKK9 pKa = 10.58QGSEE13 pKa = 4.12EE14 pKa = 4.17EE15 pKa = 4.03EE16 pKa = 4.01DD17 pKa = 3.99VYY19 pKa = 11.08GTVVFSDD26 pKa = 3.06CHH28 pKa = 6.8IVVNGNVSIVVNIDD42 pKa = 2.82AAGAEE47 pKa = 4.28EE48 pKa = 4.33QEE50 pKa = 4.62GDD52 pKa = 4.06TGDD55 pKa = 4.02DD56 pKa = 3.71TPTSDD61 pKa = 5.21GGEE64 pKa = 4.04QAQGEE69 pKa = 4.55SAEE72 pKa = 4.48AEE74 pKa = 4.04AEE76 pKa = 4.21EE77 pKa = 5.17PGDD80 pKa = 5.04SPICFQAFVPPPPPPPGPVALCCVDD105 pKa = 3.46AAQGEE110 pKa = 4.65EE111 pKa = 4.32EE112 pKa = 4.69GDD114 pKa = 3.74ANPTDD119 pKa = 4.16GADD122 pKa = 3.45EE123 pKa = 4.79DD124 pKa = 4.89GEE126 pKa = 4.38GGIDD130 pKa = 3.32VGRR133 pKa = 11.84RR134 pKa = 11.84LPDD137 pKa = 3.29GCEE140 pKa = 3.81TDD142 pKa = 3.39GGLSFEE148 pKa = 4.76AGLMVKK154 pKa = 10.18AHH156 pKa = 6.11IQRR159 pKa = 11.84SVDD162 pKa = 3.29QQDD165 pKa = 3.8SVRR168 pKa = 11.84QPSTSTQAAPAASEE182 pKa = 4.14TDD184 pKa = 3.99GPVHH188 pKa = 7.38PDD190 pKa = 2.72TGEE193 pKa = 3.95AVAPAAPAAPAAEE206 pKa = 4.7PEE208 pKa = 4.35PTPAPVPIPAPPPPAAAALTPTAAPVSPQPAAEE241 pKa = 4.51GSAPAPAAPAVV252 pKa = 3.49
MM1 pKa = 7.79ASQPKK6 pKa = 9.76RR7 pKa = 11.84FKK9 pKa = 10.58QGSEE13 pKa = 4.12EE14 pKa = 4.17EE15 pKa = 4.03EE16 pKa = 4.01DD17 pKa = 3.99VYY19 pKa = 11.08GTVVFSDD26 pKa = 3.06CHH28 pKa = 6.8IVVNGNVSIVVNIDD42 pKa = 2.82AAGAEE47 pKa = 4.28EE48 pKa = 4.33QEE50 pKa = 4.62GDD52 pKa = 4.06TGDD55 pKa = 4.02DD56 pKa = 3.71TPTSDD61 pKa = 5.21GGEE64 pKa = 4.04QAQGEE69 pKa = 4.55SAEE72 pKa = 4.48AEE74 pKa = 4.04AEE76 pKa = 4.21EE77 pKa = 5.17PGDD80 pKa = 5.04SPICFQAFVPPPPPPPGPVALCCVDD105 pKa = 3.46AAQGEE110 pKa = 4.65EE111 pKa = 4.32EE112 pKa = 4.69GDD114 pKa = 3.74ANPTDD119 pKa = 4.16GADD122 pKa = 3.45EE123 pKa = 4.79DD124 pKa = 4.89GEE126 pKa = 4.38GGIDD130 pKa = 3.32VGRR133 pKa = 11.84RR134 pKa = 11.84LPDD137 pKa = 3.29GCEE140 pKa = 3.81TDD142 pKa = 3.39GGLSFEE148 pKa = 4.76AGLMVKK154 pKa = 10.18AHH156 pKa = 6.11IQRR159 pKa = 11.84SVDD162 pKa = 3.29QQDD165 pKa = 3.8SVRR168 pKa = 11.84QPSTSTQAAPAASEE182 pKa = 4.14TDD184 pKa = 3.99GPVHH188 pKa = 7.38PDD190 pKa = 2.72TGEE193 pKa = 3.95AVAPAAPAAPAAEE206 pKa = 4.7PEE208 pKa = 4.35PTPAPVPIPAPPPPAAAALTPTAAPVSPQPAAEE241 pKa = 4.51GSAPAPAAPAVV252 pKa = 3.49
Molecular weight: 25.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A168T4T6|A0A168T4T6_9POLY ORF01L OS=Marbled eel polyomavirus OX=1662286 PE=4 SV=1
MM1 pKa = 8.01DD2 pKa = 4.54GARR5 pKa = 11.84RR6 pKa = 11.84HH7 pKa = 5.87GGGVGPWMRR16 pKa = 11.84GAGGGPYY23 pKa = 8.93WDD25 pKa = 4.37SSAAAQKK32 pKa = 9.54WLSRR36 pKa = 11.84QAGKK40 pKa = 10.54RR41 pKa = 11.84GWKK44 pKa = 8.45NRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84GGGLNIKK55 pKa = 9.73QLASKK60 pKa = 9.65AWQGIRR66 pKa = 11.84NNLIPLISRR75 pKa = 11.84HH76 pKa = 5.41ARR78 pKa = 11.84SFLSQAAEE86 pKa = 3.73HH87 pKa = 6.5AVQNGPQYY95 pKa = 11.39YY96 pKa = 10.03SAFKK100 pKa = 10.41QGGVQGLRR108 pKa = 11.84DD109 pKa = 3.85SVLDD113 pKa = 4.18NLPGMAGSFIRR124 pKa = 11.84SVAKK128 pKa = 9.54TGRR131 pKa = 11.84GSGPHH136 pKa = 6.84CATIGQMLLSARR148 pKa = 11.84PHH150 pKa = 5.66IQSVTLPMPSHH161 pKa = 6.37LRR163 pKa = 11.84SEE165 pKa = 4.64GLNRR169 pKa = 11.84AYY171 pKa = 10.74AKK173 pKa = 9.05MARR176 pKa = 11.84GEE178 pKa = 4.24SLNPYY183 pKa = 9.51SQDD186 pKa = 3.16DD187 pKa = 3.81CAAPFLAVQDD197 pKa = 3.97CALRR201 pKa = 11.84EE202 pKa = 4.35AVKK205 pKa = 10.7HH206 pKa = 5.25MNPEE210 pKa = 3.76EE211 pKa = 4.26RR212 pKa = 11.84GGFLPLLIPLLGGAISSLVGSIPKK236 pKa = 9.85FVEE239 pKa = 3.51LAQANKK245 pKa = 9.9RR246 pKa = 11.84GKK248 pKa = 10.76GSDD251 pKa = 3.67LLGMFSPPPFTSSGHH266 pKa = 4.98SPRR269 pKa = 11.84GAGPRR274 pKa = 11.84ASTFLTAQFEE284 pKa = 4.48NGATVQMSRR293 pKa = 11.84RR294 pKa = 11.84KK295 pKa = 10.04SEE297 pKa = 3.7EE298 pKa = 3.75TPYY301 pKa = 10.84KK302 pKa = 10.4IIVKK306 pKa = 10.22KK307 pKa = 10.64EE308 pKa = 3.76GGGSRR313 pKa = 11.84GRR315 pKa = 11.84PRR317 pKa = 11.84QLTWYY322 pKa = 10.4ASADD326 pKa = 3.67LFF328 pKa = 4.86
MM1 pKa = 8.01DD2 pKa = 4.54GARR5 pKa = 11.84RR6 pKa = 11.84HH7 pKa = 5.87GGGVGPWMRR16 pKa = 11.84GAGGGPYY23 pKa = 8.93WDD25 pKa = 4.37SSAAAQKK32 pKa = 9.54WLSRR36 pKa = 11.84QAGKK40 pKa = 10.54RR41 pKa = 11.84GWKK44 pKa = 8.45NRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84GGGLNIKK55 pKa = 9.73QLASKK60 pKa = 9.65AWQGIRR66 pKa = 11.84NNLIPLISRR75 pKa = 11.84HH76 pKa = 5.41ARR78 pKa = 11.84SFLSQAAEE86 pKa = 3.73HH87 pKa = 6.5AVQNGPQYY95 pKa = 11.39YY96 pKa = 10.03SAFKK100 pKa = 10.41QGGVQGLRR108 pKa = 11.84DD109 pKa = 3.85SVLDD113 pKa = 4.18NLPGMAGSFIRR124 pKa = 11.84SVAKK128 pKa = 9.54TGRR131 pKa = 11.84GSGPHH136 pKa = 6.84CATIGQMLLSARR148 pKa = 11.84PHH150 pKa = 5.66IQSVTLPMPSHH161 pKa = 6.37LRR163 pKa = 11.84SEE165 pKa = 4.64GLNRR169 pKa = 11.84AYY171 pKa = 10.74AKK173 pKa = 9.05MARR176 pKa = 11.84GEE178 pKa = 4.24SLNPYY183 pKa = 9.51SQDD186 pKa = 3.16DD187 pKa = 3.81CAAPFLAVQDD197 pKa = 3.97CALRR201 pKa = 11.84EE202 pKa = 4.35AVKK205 pKa = 10.7HH206 pKa = 5.25MNPEE210 pKa = 3.76EE211 pKa = 4.26RR212 pKa = 11.84GGFLPLLIPLLGGAISSLVGSIPKK236 pKa = 9.85FVEE239 pKa = 3.51LAQANKK245 pKa = 9.9RR246 pKa = 11.84GKK248 pKa = 10.76GSDD251 pKa = 3.67LLGMFSPPPFTSSGHH266 pKa = 4.98SPRR269 pKa = 11.84GAGPRR274 pKa = 11.84ASTFLTAQFEE284 pKa = 4.48NGATVQMSRR293 pKa = 11.84RR294 pKa = 11.84KK295 pKa = 10.04SEE297 pKa = 3.7EE298 pKa = 3.75TPYY301 pKa = 10.84KK302 pKa = 10.4IIVKK306 pKa = 10.22KK307 pKa = 10.64EE308 pKa = 3.76GGGSRR313 pKa = 11.84GRR315 pKa = 11.84PRR317 pKa = 11.84QLTWYY322 pKa = 10.4ASADD326 pKa = 3.67LFF328 pKa = 4.86
Molecular weight: 35.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4658 |
104 |
820 |
291.1 |
32.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.128 ± 0.719 | 1.889 ± 0.345 |
5.303 ± 0.621 | 5.174 ± 0.577 |
3.392 ± 0.363 | 6.977 ± 0.842 |
2.748 ± 0.281 | 4.165 ± 0.443 |
3.242 ± 0.427 | 8.437 ± 0.674 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.533 ± 0.249 | 4.122 ± 0.416 |
9.124 ± 1.3 | 4.938 ± 0.308 |
6.977 ± 0.483 | 8.416 ± 0.518 |
5.539 ± 0.488 | 6.526 ± 0.373 |
0.859 ± 0.166 | 2.512 ± 0.206 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
