
Sparus aurata (Gilthead sea bream)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha; Neoteleostei<
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
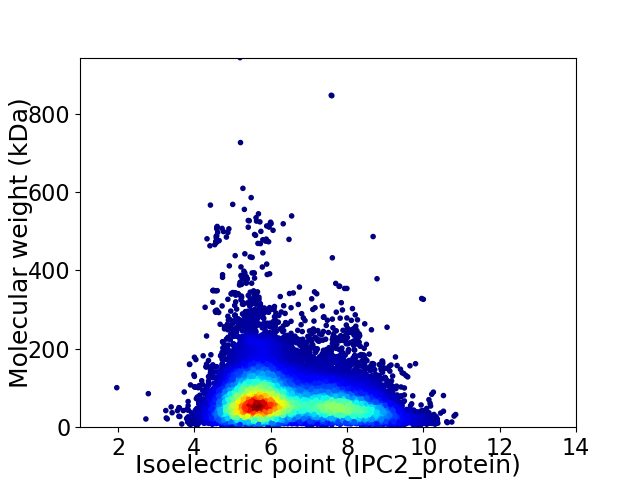
Virtual 2D-PAGE plot for 69200 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
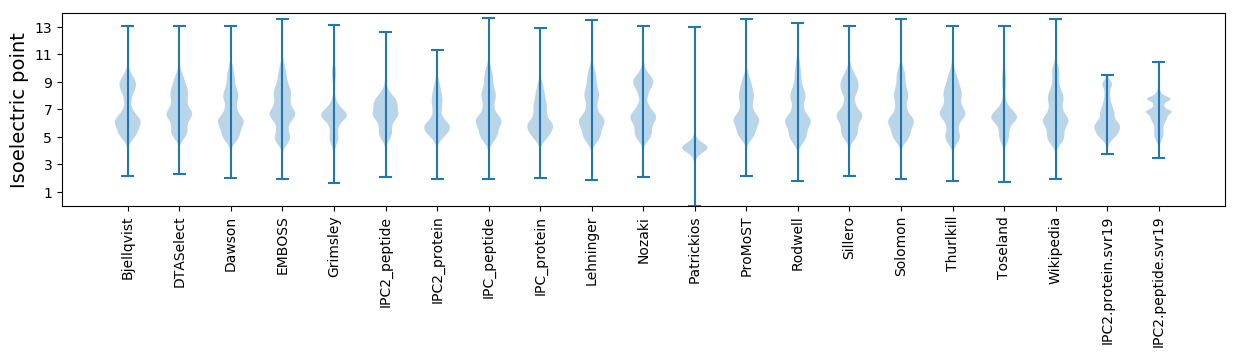
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A671VJU4|A0A671VJU4_SPAAU Isoform of A0A671VID7 BRCA1 interacting protein C-terminal helicase 1 OS=Sparus aurata OX=8175 GN=brip1 PE=4 SV=1
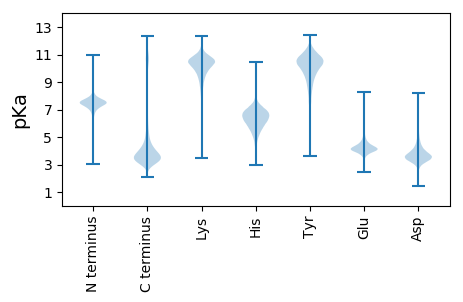
MM1 pKa = 7.6GVRR4 pKa = 11.84CINTSPGFRR13 pKa = 11.84CGSCPAGYY21 pKa = 8.41TGPQVQGVGLAYY33 pKa = 9.21ATANKK38 pKa = 9.26QVCKK42 pKa = 10.43DD43 pKa = 3.22INEE46 pKa = 4.55CSTSNGGCVEE56 pKa = 4.42NSVCMNTPGSFRR68 pKa = 11.84CGPCKK73 pKa = 9.86TGYY76 pKa = 10.67VGDD79 pKa = 3.78QRR81 pKa = 11.84RR82 pKa = 11.84GCKK85 pKa = 9.57PEE87 pKa = 3.83RR88 pKa = 11.84ACGNGQPNPCHH99 pKa = 6.71ASAEE103 pKa = 4.62CIVHH107 pKa = 6.61RR108 pKa = 11.84EE109 pKa = 3.94GKK111 pKa = 9.6IEE113 pKa = 4.14CQCGVGWAGNGYY125 pKa = 9.88LCGPDD130 pKa = 3.4SDD132 pKa = 4.6IDD134 pKa = 3.81GFPDD138 pKa = 3.68EE139 pKa = 5.77KK140 pKa = 10.84LACPDD145 pKa = 3.67TNCNKK150 pKa = 10.24DD151 pKa = 3.15NCLTVPNSGQEE162 pKa = 4.06DD163 pKa = 4.12ADD165 pKa = 3.79GDD167 pKa = 4.61GIGDD171 pKa = 4.07ACDD174 pKa = 3.53EE175 pKa = 4.74DD176 pKa = 5.69ADD178 pKa = 4.52GDD180 pKa = 4.74GILNMQDD187 pKa = 2.83NCVLVPNVDD196 pKa = 3.48QKK198 pKa = 10.85NTDD201 pKa = 3.25EE202 pKa = 5.68DD203 pKa = 4.59DD204 pKa = 5.05FGDD207 pKa = 3.87ACDD210 pKa = 3.29NCRR213 pKa = 11.84AVKK216 pKa = 10.88NNDD219 pKa = 3.3QKK221 pKa = 11.3DD222 pKa = 3.43TDD224 pKa = 3.37VDD226 pKa = 3.91KK227 pKa = 11.51FGDD230 pKa = 3.79EE231 pKa = 4.27CDD233 pKa = 3.68EE234 pKa = 5.81DD235 pKa = 4.18IDD237 pKa = 5.21GDD239 pKa = 4.45GIPNHH244 pKa = 7.08LDD246 pKa = 2.97NCKK249 pKa = 9.88RR250 pKa = 11.84VPNTDD255 pKa = 2.78QKK257 pKa = 11.7DD258 pKa = 3.53RR259 pKa = 11.84DD260 pKa = 3.6GDD262 pKa = 4.12KK263 pKa = 11.46VGDD266 pKa = 4.05ACDD269 pKa = 3.18SCPYY273 pKa = 10.08VKK275 pKa = 10.63NPEE278 pKa = 4.07QTDD281 pKa = 3.13VDD283 pKa = 3.7NDD285 pKa = 4.85LIGDD289 pKa = 3.96PCDD292 pKa = 3.62TNKK295 pKa = 10.93DD296 pKa = 3.49SDD298 pKa = 4.16GDD300 pKa = 3.81GHH302 pKa = 6.91QDD304 pKa = 4.2SRR306 pKa = 11.84DD307 pKa = 3.47NCPAVINSSQLDD319 pKa = 3.5TDD321 pKa = 3.78KK322 pKa = 11.37DD323 pKa = 4.07GKK325 pKa = 11.12GDD327 pKa = 3.71EE328 pKa = 5.3CDD330 pKa = 5.61DD331 pKa = 5.63DD332 pKa = 6.35DD333 pKa = 7.03DD334 pKa = 6.2NDD336 pKa = 5.44GIPDD340 pKa = 4.89LLPPGPDD347 pKa = 2.97NCRR350 pKa = 11.84LIPNPLQEE358 pKa = 4.62DD359 pKa = 4.3TNGDD363 pKa = 3.67GVGDD367 pKa = 3.54ICEE370 pKa = 4.37KK371 pKa = 11.1DD372 pKa = 3.39FDD374 pKa = 4.51NDD376 pKa = 4.38TIIDD380 pKa = 4.31TIDD383 pKa = 3.31VCPEE387 pKa = 3.65NAEE390 pKa = 4.04VTLTDD395 pKa = 3.59FRR397 pKa = 11.84EE398 pKa = 4.27YY399 pKa = 9.18QTVVLDD405 pKa = 4.15PEE407 pKa = 5.32GDD409 pKa = 3.65AQIDD413 pKa = 4.13PNWVVLNQYY422 pKa = 9.57IVLYY426 pKa = 9.86NSTSVTPVSPPGYY439 pKa = 7.78TAFSGVDD446 pKa = 3.72FEE448 pKa = 5.24GTFHH452 pKa = 6.81VNTVTDD458 pKa = 3.57DD459 pKa = 4.08DD460 pKa = 4.35YY461 pKa = 12.0AGFIFGYY468 pKa = 9.58QDD470 pKa = 2.83SSSFYY475 pKa = 9.91VVMWKK480 pKa = 9.52QVEE483 pKa = 4.18QIYY486 pKa = 8.33WQANPFRR493 pKa = 11.84AVAEE497 pKa = 4.08PGIQLKK503 pKa = 10.06AVKK506 pKa = 10.35SNTGPGEE513 pKa = 4.06NLRR516 pKa = 11.84NALWHH521 pKa = 6.38TGDD524 pKa = 3.08TSEE527 pKa = 5.12QVKK530 pKa = 10.14LLWKK534 pKa = 10.09DD535 pKa = 3.18ARR537 pKa = 11.84NVGWKK542 pKa = 10.41DD543 pKa = 3.0KK544 pKa = 10.83TSYY547 pKa = 10.52RR548 pKa = 11.84WFLQHH553 pKa = 6.87RR554 pKa = 11.84PADD557 pKa = 3.73GYY559 pKa = 10.45IRR561 pKa = 11.84VRR563 pKa = 11.84FYY565 pKa = 11.04EE566 pKa = 4.86GPQMVADD573 pKa = 3.78TGVIIDD579 pKa = 3.35ATMRR583 pKa = 11.84GGRR586 pKa = 11.84LGVFCFSQEE595 pKa = 3.95NIIWANLRR603 pKa = 11.84YY604 pKa = 9.71RR605 pKa = 11.84CNDD608 pKa = 3.48TLPEE612 pKa = 4.41DD613 pKa = 3.9FDD615 pKa = 3.96TYY617 pKa = 10.72RR618 pKa = 11.84AQQVQLVAA626 pKa = 4.4
MM1 pKa = 7.6GVRR4 pKa = 11.84CINTSPGFRR13 pKa = 11.84CGSCPAGYY21 pKa = 8.41TGPQVQGVGLAYY33 pKa = 9.21ATANKK38 pKa = 9.26QVCKK42 pKa = 10.43DD43 pKa = 3.22INEE46 pKa = 4.55CSTSNGGCVEE56 pKa = 4.42NSVCMNTPGSFRR68 pKa = 11.84CGPCKK73 pKa = 9.86TGYY76 pKa = 10.67VGDD79 pKa = 3.78QRR81 pKa = 11.84RR82 pKa = 11.84GCKK85 pKa = 9.57PEE87 pKa = 3.83RR88 pKa = 11.84ACGNGQPNPCHH99 pKa = 6.71ASAEE103 pKa = 4.62CIVHH107 pKa = 6.61RR108 pKa = 11.84EE109 pKa = 3.94GKK111 pKa = 9.6IEE113 pKa = 4.14CQCGVGWAGNGYY125 pKa = 9.88LCGPDD130 pKa = 3.4SDD132 pKa = 4.6IDD134 pKa = 3.81GFPDD138 pKa = 3.68EE139 pKa = 5.77KK140 pKa = 10.84LACPDD145 pKa = 3.67TNCNKK150 pKa = 10.24DD151 pKa = 3.15NCLTVPNSGQEE162 pKa = 4.06DD163 pKa = 4.12ADD165 pKa = 3.79GDD167 pKa = 4.61GIGDD171 pKa = 4.07ACDD174 pKa = 3.53EE175 pKa = 4.74DD176 pKa = 5.69ADD178 pKa = 4.52GDD180 pKa = 4.74GILNMQDD187 pKa = 2.83NCVLVPNVDD196 pKa = 3.48QKK198 pKa = 10.85NTDD201 pKa = 3.25EE202 pKa = 5.68DD203 pKa = 4.59DD204 pKa = 5.05FGDD207 pKa = 3.87ACDD210 pKa = 3.29NCRR213 pKa = 11.84AVKK216 pKa = 10.88NNDD219 pKa = 3.3QKK221 pKa = 11.3DD222 pKa = 3.43TDD224 pKa = 3.37VDD226 pKa = 3.91KK227 pKa = 11.51FGDD230 pKa = 3.79EE231 pKa = 4.27CDD233 pKa = 3.68EE234 pKa = 5.81DD235 pKa = 4.18IDD237 pKa = 5.21GDD239 pKa = 4.45GIPNHH244 pKa = 7.08LDD246 pKa = 2.97NCKK249 pKa = 9.88RR250 pKa = 11.84VPNTDD255 pKa = 2.78QKK257 pKa = 11.7DD258 pKa = 3.53RR259 pKa = 11.84DD260 pKa = 3.6GDD262 pKa = 4.12KK263 pKa = 11.46VGDD266 pKa = 4.05ACDD269 pKa = 3.18SCPYY273 pKa = 10.08VKK275 pKa = 10.63NPEE278 pKa = 4.07QTDD281 pKa = 3.13VDD283 pKa = 3.7NDD285 pKa = 4.85LIGDD289 pKa = 3.96PCDD292 pKa = 3.62TNKK295 pKa = 10.93DD296 pKa = 3.49SDD298 pKa = 4.16GDD300 pKa = 3.81GHH302 pKa = 6.91QDD304 pKa = 4.2SRR306 pKa = 11.84DD307 pKa = 3.47NCPAVINSSQLDD319 pKa = 3.5TDD321 pKa = 3.78KK322 pKa = 11.37DD323 pKa = 4.07GKK325 pKa = 11.12GDD327 pKa = 3.71EE328 pKa = 5.3CDD330 pKa = 5.61DD331 pKa = 5.63DD332 pKa = 6.35DD333 pKa = 7.03DD334 pKa = 6.2NDD336 pKa = 5.44GIPDD340 pKa = 4.89LLPPGPDD347 pKa = 2.97NCRR350 pKa = 11.84LIPNPLQEE358 pKa = 4.62DD359 pKa = 4.3TNGDD363 pKa = 3.67GVGDD367 pKa = 3.54ICEE370 pKa = 4.37KK371 pKa = 11.1DD372 pKa = 3.39FDD374 pKa = 4.51NDD376 pKa = 4.38TIIDD380 pKa = 4.31TIDD383 pKa = 3.31VCPEE387 pKa = 3.65NAEE390 pKa = 4.04VTLTDD395 pKa = 3.59FRR397 pKa = 11.84EE398 pKa = 4.27YY399 pKa = 9.18QTVVLDD405 pKa = 4.15PEE407 pKa = 5.32GDD409 pKa = 3.65AQIDD413 pKa = 4.13PNWVVLNQYY422 pKa = 9.57IVLYY426 pKa = 9.86NSTSVTPVSPPGYY439 pKa = 7.78TAFSGVDD446 pKa = 3.72FEE448 pKa = 5.24GTFHH452 pKa = 6.81VNTVTDD458 pKa = 3.57DD459 pKa = 4.08DD460 pKa = 4.35YY461 pKa = 12.0AGFIFGYY468 pKa = 9.58QDD470 pKa = 2.83SSSFYY475 pKa = 9.91VVMWKK480 pKa = 9.52QVEE483 pKa = 4.18QIYY486 pKa = 8.33WQANPFRR493 pKa = 11.84AVAEE497 pKa = 4.08PGIQLKK503 pKa = 10.06AVKK506 pKa = 10.35SNTGPGEE513 pKa = 4.06NLRR516 pKa = 11.84NALWHH521 pKa = 6.38TGDD524 pKa = 3.08TSEE527 pKa = 5.12QVKK530 pKa = 10.14LLWKK534 pKa = 10.09DD535 pKa = 3.18ARR537 pKa = 11.84NVGWKK542 pKa = 10.41DD543 pKa = 3.0KK544 pKa = 10.83TSYY547 pKa = 10.52RR548 pKa = 11.84WFLQHH553 pKa = 6.87RR554 pKa = 11.84PADD557 pKa = 3.73GYY559 pKa = 10.45IRR561 pKa = 11.84VRR563 pKa = 11.84FYY565 pKa = 11.04EE566 pKa = 4.86GPQMVADD573 pKa = 3.78TGVIIDD579 pKa = 3.35ATMRR583 pKa = 11.84GGRR586 pKa = 11.84LGVFCFSQEE595 pKa = 3.95NIIWANLRR603 pKa = 11.84YY604 pKa = 9.71RR605 pKa = 11.84CNDD608 pKa = 3.48TLPEE612 pKa = 4.41DD613 pKa = 3.9FDD615 pKa = 3.96TYY617 pKa = 10.72RR618 pKa = 11.84AQQVQLVAA626 pKa = 4.4
Molecular weight: 68.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A671UXB6|A0A671UXB6_SPAAU 2 4-dienoyl-CoA reductase 1 OS=Sparus aurata OX=8175 GN=decr1 PE=4 SV=1
MM1 pKa = 6.64WTPGIVWTPRR11 pKa = 11.84IVIVWTPGIMWTLRR25 pKa = 11.84IVWTPGIVWTPGIMWTPRR43 pKa = 11.84IVWTPRR49 pKa = 11.84IMWTPGIVWTPGIMWTPGIVWTPRR73 pKa = 11.84IVWTPGIMWTLRR85 pKa = 11.84IVWTPGIVWTPGIMWTPRR103 pKa = 11.84IVWTPRR109 pKa = 11.84IMWTPGIVWTPGIMWIPGIVWTPRR133 pKa = 11.84IVVTPGIVGTPGIVWTPGIMWTPGIVWTPGIMWTPRR169 pKa = 11.84IVWTPGIMWTLRR181 pKa = 11.84IVWTPGIVWTPGIMWTLRR199 pKa = 11.84IVWTPGIVWTPGIMWTTRR217 pKa = 11.84IVGTPGIVGTPGIVWTPGIMWTPGIVWTPGIMWTPRR253 pKa = 11.84IVWTPGIVWTPGIMWTPGIVWTPGIMWTPRR283 pKa = 11.84IVWTPGIMWALRR295 pKa = 11.84IGWTPGIVWTPGIMWTLRR313 pKa = 11.84IVWTPGIVWTPGIMWTTRR331 pKa = 11.84IVGTPGIVGTPGIMWTPRR349 pKa = 11.84IVIGWTPGIVWTPGIMWTLRR369 pKa = 11.84IVWTPGIVWTPGIMWTTRR387 pKa = 11.84IMWTPGIVWTPGIMWTPRR405 pKa = 11.84IVWTPGIVWTPGIMWTPGIVWTPGIMWTPRR435 pKa = 11.84IVWTPGIMWTLRR447 pKa = 11.84IVWTPGIVWTPGIMWTLRR465 pKa = 11.84IVWTPGIVWTPGIMWTTRR483 pKa = 11.84IVGTPGIVGTPGIMWTPGIVWTPGIMWTPRR513 pKa = 11.84IVWTPGIVWTPGIMWTPGIVWTPGIMWTPRR543 pKa = 11.84IVWTPGIMWTLRR555 pKa = 11.84IVWTPGIVWTPRR567 pKa = 11.84IVWTPGIMWTPRR579 pKa = 11.84IVWTPGIMWTLRR591 pKa = 11.84IVWTPGIVWTPGIMWTLRR609 pKa = 11.84IVWTPGIVWTPGIMWTTRR627 pKa = 11.84IVGTPGIVGTPGIVWTPGIMWTPRR651 pKa = 11.84IVWTPGIMWTPRR663 pKa = 11.84IVWTPGIVWTPGIMWTPGIVWTPGIMWTPRR693 pKa = 11.84IVWTPGIMWTLRR705 pKa = 11.84IVWTPGIVWTPGIMWTLRR723 pKa = 11.84IVWTPGIVWTPGIMWTTRR741 pKa = 11.84IVGTPGIVGTPGIVVTPGIVWTPRR765 pKa = 11.84IVGLVCQHH773 pKa = 6.69SISSAIVLKK782 pKa = 10.67GHH784 pKa = 7.2LPGLVTGPTEE794 pKa = 4.17FMNSFHH800 pKa = 6.93SS801 pKa = 3.7
MM1 pKa = 6.64WTPGIVWTPRR11 pKa = 11.84IVIVWTPGIMWTLRR25 pKa = 11.84IVWTPGIVWTPGIMWTPRR43 pKa = 11.84IVWTPRR49 pKa = 11.84IMWTPGIVWTPGIMWTPGIVWTPRR73 pKa = 11.84IVWTPGIMWTLRR85 pKa = 11.84IVWTPGIVWTPGIMWTPRR103 pKa = 11.84IVWTPRR109 pKa = 11.84IMWTPGIVWTPGIMWIPGIVWTPRR133 pKa = 11.84IVVTPGIVGTPGIVWTPGIMWTPGIVWTPGIMWTPRR169 pKa = 11.84IVWTPGIMWTLRR181 pKa = 11.84IVWTPGIVWTPGIMWTLRR199 pKa = 11.84IVWTPGIVWTPGIMWTTRR217 pKa = 11.84IVGTPGIVGTPGIVWTPGIMWTPGIVWTPGIMWTPRR253 pKa = 11.84IVWTPGIVWTPGIMWTPGIVWTPGIMWTPRR283 pKa = 11.84IVWTPGIMWALRR295 pKa = 11.84IGWTPGIVWTPGIMWTLRR313 pKa = 11.84IVWTPGIVWTPGIMWTTRR331 pKa = 11.84IVGTPGIVGTPGIMWTPRR349 pKa = 11.84IVIGWTPGIVWTPGIMWTLRR369 pKa = 11.84IVWTPGIVWTPGIMWTTRR387 pKa = 11.84IMWTPGIVWTPGIMWTPRR405 pKa = 11.84IVWTPGIVWTPGIMWTPGIVWTPGIMWTPRR435 pKa = 11.84IVWTPGIMWTLRR447 pKa = 11.84IVWTPGIVWTPGIMWTLRR465 pKa = 11.84IVWTPGIVWTPGIMWTTRR483 pKa = 11.84IVGTPGIVGTPGIMWTPGIVWTPGIMWTPRR513 pKa = 11.84IVWTPGIVWTPGIMWTPGIVWTPGIMWTPRR543 pKa = 11.84IVWTPGIMWTLRR555 pKa = 11.84IVWTPGIVWTPRR567 pKa = 11.84IVWTPGIMWTPRR579 pKa = 11.84IVWTPGIMWTLRR591 pKa = 11.84IVWTPGIVWTPGIMWTLRR609 pKa = 11.84IVWTPGIVWTPGIMWTTRR627 pKa = 11.84IVGTPGIVGTPGIVWTPGIMWTPRR651 pKa = 11.84IVWTPGIMWTPRR663 pKa = 11.84IVWTPGIVWTPGIMWTPGIVWTPGIMWTPRR693 pKa = 11.84IVWTPGIMWTLRR705 pKa = 11.84IVWTPGIVWTPGIMWTLRR723 pKa = 11.84IVWTPGIVWTPGIMWTTRR741 pKa = 11.84IVGTPGIVGTPGIVVTPGIVWTPRR765 pKa = 11.84IVGLVCQHH773 pKa = 6.69SISSAIVLKK782 pKa = 10.67GHH784 pKa = 7.2LPGLVTGPTEE794 pKa = 4.17FMNSFHH800 pKa = 6.93SS801 pKa = 3.7
Molecular weight: 91.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
47700975 |
16 |
8613 |
689.3 |
77.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.481 ± 0.007 | 2.299 ± 0.007 |
5.281 ± 0.006 | 6.808 ± 0.011 |
3.776 ± 0.006 | 6.279 ± 0.009 |
2.629 ± 0.004 | 4.595 ± 0.006 |
5.605 ± 0.01 | 9.653 ± 0.01 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.405 ± 0.004 | 3.899 ± 0.004 |
5.454 ± 0.009 | 4.659 ± 0.007 |
5.577 ± 0.007 | 8.357 ± 0.009 |
5.685 ± 0.009 | 6.496 ± 0.006 |
1.184 ± 0.003 | 2.87 ± 0.005 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
