
Providence virus (isolate Helicoverpa zea/United States/vFLM1/-) (PRVFL)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Carmotetraviridae; Alphacarmotetravirus; Providence virus
Average proteome isoelectric point is 7.68
Get precalculated fractions of proteins

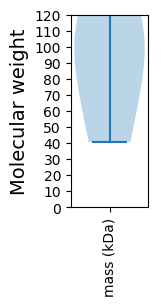
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
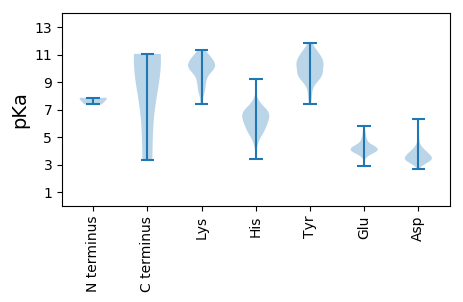
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q80IX5|Q80IX5_PRVFL p81 OS=Providence virus (isolate Helicoverpa zea/United States/vFLM1/-) OX=1289469 PE=1 SV=1
MM1 pKa = 7.37QNLPVPNGEE10 pKa = 4.65SPPKK14 pKa = 8.88TDD16 pKa = 2.84EE17 pKa = 4.06VRR19 pKa = 11.84DD20 pKa = 3.69SRR22 pKa = 11.84RR23 pKa = 11.84NSDD26 pKa = 3.81DD27 pKa = 3.85EE28 pKa = 4.15EE29 pKa = 5.41PEE31 pKa = 4.13YY32 pKa = 10.67PRR34 pKa = 11.84GDD36 pKa = 4.23PIEE39 pKa = 5.5DD40 pKa = 3.59LTDD43 pKa = 4.3DD44 pKa = 4.72GDD46 pKa = 3.81IEE48 pKa = 4.72KK49 pKa = 11.07NPGPASANQAGKK61 pKa = 10.31RR62 pKa = 11.84LEE64 pKa = 3.85PRR66 pKa = 11.84KK67 pKa = 8.37RR68 pKa = 11.84TRR70 pKa = 11.84SIRR73 pKa = 11.84SRR75 pKa = 11.84QDD77 pKa = 3.18LDD79 pKa = 3.28TMHH82 pKa = 6.88RR83 pKa = 11.84VVVSVDD89 pKa = 3.03VDD91 pKa = 3.82SLTTLMGNIMTLAGSGGRR109 pKa = 11.84GSLLTAGDD117 pKa = 3.84VEE119 pKa = 4.97KK120 pKa = 11.35NPGPQMTRR128 pKa = 11.84EE129 pKa = 3.84NKK131 pKa = 9.57APNMSVRR138 pKa = 11.84AQNVTVSNSRR148 pKa = 11.84RR149 pKa = 11.84RR150 pKa = 11.84RR151 pKa = 11.84RR152 pKa = 11.84KK153 pKa = 8.25NRR155 pKa = 11.84KK156 pKa = 7.54PRR158 pKa = 11.84KK159 pKa = 8.38QVEE162 pKa = 3.88QATILKK168 pKa = 8.76PQLLPGEE175 pKa = 4.5SVAPSGGRR183 pKa = 11.84GTMDD187 pKa = 3.57PPVHH191 pKa = 6.63EE192 pKa = 5.12ICAQSIDD199 pKa = 3.88PSEE202 pKa = 4.37GAVGWFYY209 pKa = 11.64KK210 pKa = 11.03YY211 pKa = 9.53MDD213 pKa = 4.17PAGAVEE219 pKa = 4.25SGKK222 pKa = 10.73ALGEE226 pKa = 3.96FSKK229 pKa = 11.34VPDD232 pKa = 3.53GLLRR236 pKa = 11.84YY237 pKa = 9.91SVDD240 pKa = 3.15AEE242 pKa = 3.78QRR244 pKa = 11.84PIVTIEE250 pKa = 4.18CPTVSEE256 pKa = 4.69SDD258 pKa = 3.65LPLDD262 pKa = 3.88GKK264 pKa = 10.02LWRR267 pKa = 11.84VSFISFPAFRR277 pKa = 11.84LNFIALANINNEE289 pKa = 3.79ALTLEE294 pKa = 4.27TRR296 pKa = 11.84NTFIQTLNNITNWRR310 pKa = 11.84DD311 pKa = 3.0LGTGQWAQFAPGWYY325 pKa = 8.96YY326 pKa = 10.96SIYY329 pKa = 10.55VLPNTYY335 pKa = 11.18AMAEE339 pKa = 3.87IGDD342 pKa = 4.01RR343 pKa = 11.84TDD345 pKa = 3.43SVTQFRR351 pKa = 11.84KK352 pKa = 9.02VYY354 pKa = 10.37KK355 pKa = 10.72GITFEE360 pKa = 4.91FNAPTLIDD368 pKa = 3.42QGWWVGAHH376 pKa = 6.22IPVKK380 pKa = 9.47PQSEE384 pKa = 4.68TIPAAEE390 pKa = 4.17RR391 pKa = 11.84FSAGSMTVSASNAIFQPSNTVARR414 pKa = 11.84IVWSITPLPVATVALTTGTGGTNNTSGKK442 pKa = 9.34FFSVEE447 pKa = 2.89IDD449 pKa = 3.62GNVNSVWTFTAPASILAEE467 pKa = 3.9GEE469 pKa = 4.09PFAEE473 pKa = 4.67EE474 pKa = 4.32GDD476 pKa = 3.86TTSFSMTTITADD488 pKa = 3.4TVVYY492 pKa = 10.14SVSSSLTGSSVIVRR506 pKa = 11.84GVTKK510 pKa = 10.64GSGVSITPVTVGIDD524 pKa = 3.42TEE526 pKa = 4.4AVNRR530 pKa = 11.84LSIEE534 pKa = 4.1MPALTTEE541 pKa = 4.34EE542 pKa = 4.27VTTNVPKK549 pKa = 10.94YY550 pKa = 9.86EE551 pKa = 3.92QFLCKK556 pKa = 10.06EE557 pKa = 4.0SGGAYY562 pKa = 8.28IVHH565 pKa = 6.29YY566 pKa = 10.69KK567 pKa = 9.43MNNPVFEE574 pKa = 4.34MTGEE578 pKa = 4.05EE579 pKa = 4.14NFGGFQFHH587 pKa = 6.58YY588 pKa = 10.08PGYY591 pKa = 10.35DD592 pKa = 3.35PEE594 pKa = 4.94NNALGLRR601 pKa = 11.84GIVDD605 pKa = 3.41TFEE608 pKa = 5.34NNFSSAVVHH617 pKa = 6.46FWGISQSATIVCKK630 pKa = 10.02TYY632 pKa = 10.82DD633 pKa = 3.23GWEE636 pKa = 4.15GTTNAGSTVGQFAHH650 pKa = 6.62TGAEE654 pKa = 4.09EE655 pKa = 3.88EE656 pKa = 4.66DD657 pKa = 4.07EE658 pKa = 4.58VVQLANRR665 pKa = 11.84LQMEE669 pKa = 4.59LTGVYY674 pKa = 9.74QADD677 pKa = 3.83DD678 pKa = 3.77NFAGTVSALASIGLGLLGKK697 pKa = 10.38SSATPSVIKK706 pKa = 10.73GIAQQAVGAVQANPGILEE724 pKa = 4.26GAVKK728 pKa = 10.79AIGSVGARR736 pKa = 11.84LVGSIKK742 pKa = 10.27ARR744 pKa = 11.84RR745 pKa = 11.84ARR747 pKa = 11.84RR748 pKa = 11.84RR749 pKa = 11.84ARR751 pKa = 11.84RR752 pKa = 11.84AKK754 pKa = 10.36
MM1 pKa = 7.37QNLPVPNGEE10 pKa = 4.65SPPKK14 pKa = 8.88TDD16 pKa = 2.84EE17 pKa = 4.06VRR19 pKa = 11.84DD20 pKa = 3.69SRR22 pKa = 11.84RR23 pKa = 11.84NSDD26 pKa = 3.81DD27 pKa = 3.85EE28 pKa = 4.15EE29 pKa = 5.41PEE31 pKa = 4.13YY32 pKa = 10.67PRR34 pKa = 11.84GDD36 pKa = 4.23PIEE39 pKa = 5.5DD40 pKa = 3.59LTDD43 pKa = 4.3DD44 pKa = 4.72GDD46 pKa = 3.81IEE48 pKa = 4.72KK49 pKa = 11.07NPGPASANQAGKK61 pKa = 10.31RR62 pKa = 11.84LEE64 pKa = 3.85PRR66 pKa = 11.84KK67 pKa = 8.37RR68 pKa = 11.84TRR70 pKa = 11.84SIRR73 pKa = 11.84SRR75 pKa = 11.84QDD77 pKa = 3.18LDD79 pKa = 3.28TMHH82 pKa = 6.88RR83 pKa = 11.84VVVSVDD89 pKa = 3.03VDD91 pKa = 3.82SLTTLMGNIMTLAGSGGRR109 pKa = 11.84GSLLTAGDD117 pKa = 3.84VEE119 pKa = 4.97KK120 pKa = 11.35NPGPQMTRR128 pKa = 11.84EE129 pKa = 3.84NKK131 pKa = 9.57APNMSVRR138 pKa = 11.84AQNVTVSNSRR148 pKa = 11.84RR149 pKa = 11.84RR150 pKa = 11.84RR151 pKa = 11.84RR152 pKa = 11.84KK153 pKa = 8.25NRR155 pKa = 11.84KK156 pKa = 7.54PRR158 pKa = 11.84KK159 pKa = 8.38QVEE162 pKa = 3.88QATILKK168 pKa = 8.76PQLLPGEE175 pKa = 4.5SVAPSGGRR183 pKa = 11.84GTMDD187 pKa = 3.57PPVHH191 pKa = 6.63EE192 pKa = 5.12ICAQSIDD199 pKa = 3.88PSEE202 pKa = 4.37GAVGWFYY209 pKa = 11.64KK210 pKa = 11.03YY211 pKa = 9.53MDD213 pKa = 4.17PAGAVEE219 pKa = 4.25SGKK222 pKa = 10.73ALGEE226 pKa = 3.96FSKK229 pKa = 11.34VPDD232 pKa = 3.53GLLRR236 pKa = 11.84YY237 pKa = 9.91SVDD240 pKa = 3.15AEE242 pKa = 3.78QRR244 pKa = 11.84PIVTIEE250 pKa = 4.18CPTVSEE256 pKa = 4.69SDD258 pKa = 3.65LPLDD262 pKa = 3.88GKK264 pKa = 10.02LWRR267 pKa = 11.84VSFISFPAFRR277 pKa = 11.84LNFIALANINNEE289 pKa = 3.79ALTLEE294 pKa = 4.27TRR296 pKa = 11.84NTFIQTLNNITNWRR310 pKa = 11.84DD311 pKa = 3.0LGTGQWAQFAPGWYY325 pKa = 8.96YY326 pKa = 10.96SIYY329 pKa = 10.55VLPNTYY335 pKa = 11.18AMAEE339 pKa = 3.87IGDD342 pKa = 4.01RR343 pKa = 11.84TDD345 pKa = 3.43SVTQFRR351 pKa = 11.84KK352 pKa = 9.02VYY354 pKa = 10.37KK355 pKa = 10.72GITFEE360 pKa = 4.91FNAPTLIDD368 pKa = 3.42QGWWVGAHH376 pKa = 6.22IPVKK380 pKa = 9.47PQSEE384 pKa = 4.68TIPAAEE390 pKa = 4.17RR391 pKa = 11.84FSAGSMTVSASNAIFQPSNTVARR414 pKa = 11.84IVWSITPLPVATVALTTGTGGTNNTSGKK442 pKa = 9.34FFSVEE447 pKa = 2.89IDD449 pKa = 3.62GNVNSVWTFTAPASILAEE467 pKa = 3.9GEE469 pKa = 4.09PFAEE473 pKa = 4.67EE474 pKa = 4.32GDD476 pKa = 3.86TTSFSMTTITADD488 pKa = 3.4TVVYY492 pKa = 10.14SVSSSLTGSSVIVRR506 pKa = 11.84GVTKK510 pKa = 10.64GSGVSITPVTVGIDD524 pKa = 3.42TEE526 pKa = 4.4AVNRR530 pKa = 11.84LSIEE534 pKa = 4.1MPALTTEE541 pKa = 4.34EE542 pKa = 4.27VTTNVPKK549 pKa = 10.94YY550 pKa = 9.86EE551 pKa = 3.92QFLCKK556 pKa = 10.06EE557 pKa = 4.0SGGAYY562 pKa = 8.28IVHH565 pKa = 6.29YY566 pKa = 10.69KK567 pKa = 9.43MNNPVFEE574 pKa = 4.34MTGEE578 pKa = 4.05EE579 pKa = 4.14NFGGFQFHH587 pKa = 6.58YY588 pKa = 10.08PGYY591 pKa = 10.35DD592 pKa = 3.35PEE594 pKa = 4.94NNALGLRR601 pKa = 11.84GIVDD605 pKa = 3.41TFEE608 pKa = 5.34NNFSSAVVHH617 pKa = 6.46FWGISQSATIVCKK630 pKa = 10.02TYY632 pKa = 10.82DD633 pKa = 3.23GWEE636 pKa = 4.15GTTNAGSTVGQFAHH650 pKa = 6.62TGAEE654 pKa = 4.09EE655 pKa = 3.88EE656 pKa = 4.66DD657 pKa = 4.07EE658 pKa = 4.58VVQLANRR665 pKa = 11.84LQMEE669 pKa = 4.59LTGVYY674 pKa = 9.74QADD677 pKa = 3.83DD678 pKa = 3.77NFAGTVSALASIGLGLLGKK697 pKa = 10.38SSATPSVIKK706 pKa = 10.73GIAQQAVGAVQANPGILEE724 pKa = 4.26GAVKK728 pKa = 10.79AIGSVGARR736 pKa = 11.84LVGSIKK742 pKa = 10.27ARR744 pKa = 11.84RR745 pKa = 11.84ARR747 pKa = 11.84RR748 pKa = 11.84RR749 pKa = 11.84ARR751 pKa = 11.84RR752 pKa = 11.84AKK754 pKa = 10.36
Molecular weight: 81.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q80IX5|Q80IX5_PRVFL p81 OS=Providence virus (isolate Helicoverpa zea/United States/vFLM1/-) OX=1289469 PE=1 SV=1
MM1 pKa = 7.85CEE3 pKa = 3.99PLSLRR8 pKa = 11.84LCISCKK14 pKa = 9.77ATTSSSEE21 pKa = 3.97SRR23 pKa = 11.84GEE25 pKa = 3.9EE26 pKa = 3.88LLSGSKK32 pKa = 10.17SPPPPKK38 pKa = 9.96PSLTQTLLRR47 pKa = 11.84YY48 pKa = 9.33LSSMLTHH55 pKa = 7.02LGAAIMSPLSVVTGRR70 pKa = 11.84IIPSGNTILRR80 pKa = 11.84ASLALTSATCFYY92 pKa = 10.22THH94 pKa = 6.65LRR96 pKa = 11.84GYY98 pKa = 10.47RR99 pKa = 11.84YY100 pKa = 9.53ILRR103 pKa = 11.84FCRR106 pKa = 11.84TDD108 pKa = 3.45LVLFTQSVLNLLLQRR123 pKa = 11.84IGLKK127 pKa = 8.89SLRR130 pKa = 11.84LSLDD134 pKa = 3.32TTISLPFVIPQFVNIVDD151 pKa = 3.85YY152 pKa = 11.14AQLSQALQHH161 pKa = 5.98TRR163 pKa = 11.84THH165 pKa = 5.48TRR167 pKa = 11.84SATFLNVSHH176 pKa = 7.26IPDD179 pKa = 3.2QLLRR183 pKa = 11.84FAEE186 pKa = 4.4KK187 pKa = 10.23SWIRR191 pKa = 11.84QQLRR195 pKa = 11.84HH196 pKa = 5.74VDD198 pKa = 3.43LMGPAEE204 pKa = 4.25VVEE207 pKa = 4.74FQHH210 pKa = 6.14WLSNRR215 pKa = 11.84LVDD218 pKa = 6.29LYY220 pKa = 9.47PTWRR224 pKa = 11.84PLLAALGLIALAWCLTPVARR244 pKa = 11.84VLLEE248 pKa = 4.05EE249 pKa = 5.05GEE251 pKa = 4.39EE252 pKa = 3.97KK253 pKa = 11.11DD254 pKa = 3.46PVVRR258 pKa = 11.84NLTAVLRR265 pKa = 11.84SHH267 pKa = 7.66AIFCHH272 pKa = 5.23RR273 pKa = 11.84TAGTRR278 pKa = 11.84RR279 pKa = 11.84MLADD283 pKa = 4.24RR284 pKa = 11.84ASAWLRR290 pKa = 11.84KK291 pKa = 7.46EE292 pKa = 3.98HH293 pKa = 6.87PNMEE297 pKa = 4.27EE298 pKa = 3.77HH299 pKa = 6.4MKK301 pKa = 9.9PALVARR307 pKa = 11.84AVALAMIPCRR317 pKa = 11.84EE318 pKa = 4.09EE319 pKa = 3.85VEE321 pKa = 4.81SLIDD325 pKa = 3.54QQKK328 pKa = 10.7NSASLWWVTNLLRR341 pKa = 11.84SGWNSPWEE349 pKa = 3.96WLLGRR354 pKa = 11.84TIPTKK359 pKa = 10.72
MM1 pKa = 7.85CEE3 pKa = 3.99PLSLRR8 pKa = 11.84LCISCKK14 pKa = 9.77ATTSSSEE21 pKa = 3.97SRR23 pKa = 11.84GEE25 pKa = 3.9EE26 pKa = 3.88LLSGSKK32 pKa = 10.17SPPPPKK38 pKa = 9.96PSLTQTLLRR47 pKa = 11.84YY48 pKa = 9.33LSSMLTHH55 pKa = 7.02LGAAIMSPLSVVTGRR70 pKa = 11.84IIPSGNTILRR80 pKa = 11.84ASLALTSATCFYY92 pKa = 10.22THH94 pKa = 6.65LRR96 pKa = 11.84GYY98 pKa = 10.47RR99 pKa = 11.84YY100 pKa = 9.53ILRR103 pKa = 11.84FCRR106 pKa = 11.84TDD108 pKa = 3.45LVLFTQSVLNLLLQRR123 pKa = 11.84IGLKK127 pKa = 8.89SLRR130 pKa = 11.84LSLDD134 pKa = 3.32TTISLPFVIPQFVNIVDD151 pKa = 3.85YY152 pKa = 11.14AQLSQALQHH161 pKa = 5.98TRR163 pKa = 11.84THH165 pKa = 5.48TRR167 pKa = 11.84SATFLNVSHH176 pKa = 7.26IPDD179 pKa = 3.2QLLRR183 pKa = 11.84FAEE186 pKa = 4.4KK187 pKa = 10.23SWIRR191 pKa = 11.84QQLRR195 pKa = 11.84HH196 pKa = 5.74VDD198 pKa = 3.43LMGPAEE204 pKa = 4.25VVEE207 pKa = 4.74FQHH210 pKa = 6.14WLSNRR215 pKa = 11.84LVDD218 pKa = 6.29LYY220 pKa = 9.47PTWRR224 pKa = 11.84PLLAALGLIALAWCLTPVARR244 pKa = 11.84VLLEE248 pKa = 4.05EE249 pKa = 5.05GEE251 pKa = 4.39EE252 pKa = 3.97KK253 pKa = 11.11DD254 pKa = 3.46PVVRR258 pKa = 11.84NLTAVLRR265 pKa = 11.84SHH267 pKa = 7.66AIFCHH272 pKa = 5.23RR273 pKa = 11.84TAGTRR278 pKa = 11.84RR279 pKa = 11.84MLADD283 pKa = 4.24RR284 pKa = 11.84ASAWLRR290 pKa = 11.84KK291 pKa = 7.46EE292 pKa = 3.98HH293 pKa = 6.87PNMEE297 pKa = 4.27EE298 pKa = 3.77HH299 pKa = 6.4MKK301 pKa = 9.9PALVARR307 pKa = 11.84AVALAMIPCRR317 pKa = 11.84EE318 pKa = 4.09EE319 pKa = 3.85VEE321 pKa = 4.81SLIDD325 pKa = 3.54QQKK328 pKa = 10.7NSASLWWVTNLLRR341 pKa = 11.84SGWNSPWEE349 pKa = 3.96WLLGRR354 pKa = 11.84TIPTKK359 pKa = 10.72
Molecular weight: 40.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3248 |
359 |
1220 |
812.0 |
89.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.22 ± 0.599 | 1.817 ± 0.341 |
3.972 ± 0.229 | 5.45 ± 0.242 |
2.494 ± 0.375 | 7.512 ± 1.601 |
2.679 ± 0.479 | 3.941 ± 0.703 |
3.725 ± 0.131 | 9.483 ± 2.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.97 ± 0.573 | 3.756 ± 0.45 |
6.435 ± 0.206 | 4.126 ± 0.195 |
7.112 ± 0.363 | 8.621 ± 0.429 |
6.866 ± 0.437 | 7.574 ± 0.434 |
1.817 ± 0.425 | 2.401 ± 0.209 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
