
Phialophora americana
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Chaetothyriomycetidae; Chaetothyriales; Herpotrichiellaceae; Phialophora
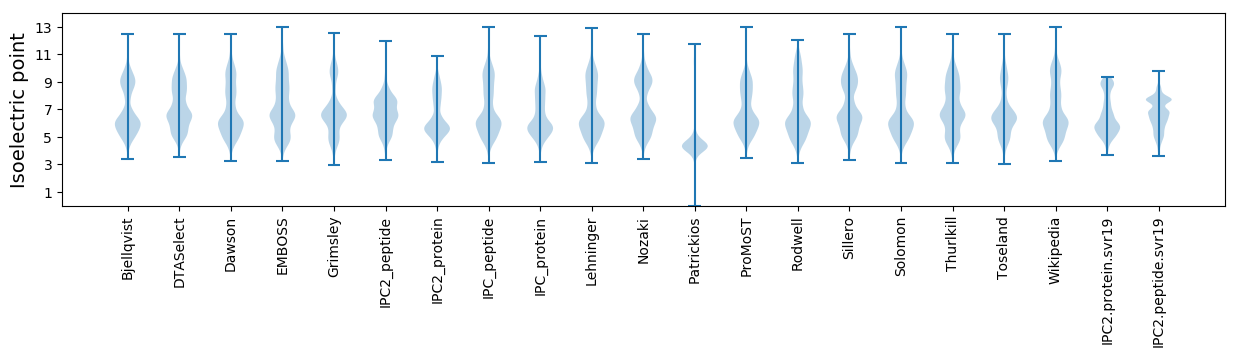
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
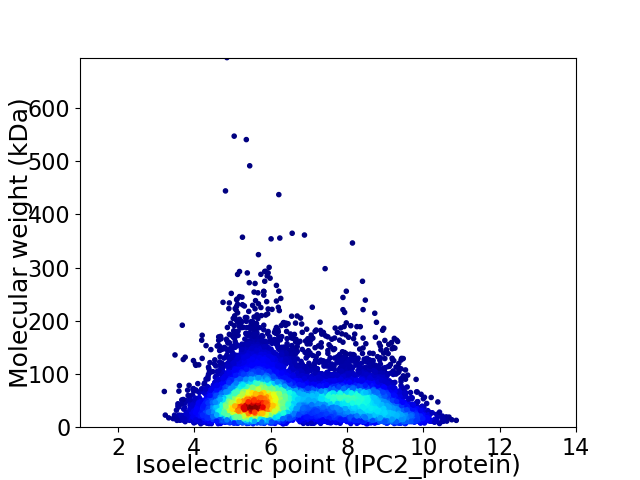
Virtual 2D-PAGE plot for 11503 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D2FUH5|A0A0D2FUH5_9EURO RNA helicase OS=Phialophora americana OX=5601 GN=PV04_08707 PE=4 SV=1
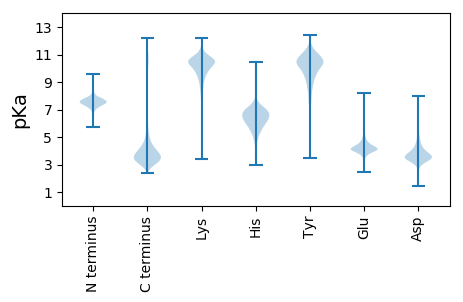
MM1 pKa = 7.16KK2 pKa = 10.19RR3 pKa = 11.84STRR6 pKa = 11.84AMPVAALLMAAANAQTTIINDD27 pKa = 3.87PADD30 pKa = 4.33PDD32 pKa = 3.65ATTVYY37 pKa = 10.99YY38 pKa = 7.95STCPTALTTTTTLSSTITFTCPGGEE63 pKa = 4.06CSGPTITPPPHH74 pKa = 6.53GSAQVSEE81 pKa = 4.21ILAFTTTLPDD91 pKa = 3.41GSVQEE96 pKa = 3.75IHH98 pKa = 6.95EE99 pKa = 4.96YY100 pKa = 8.35ITMYY104 pKa = 9.76PAIVSDD110 pKa = 4.46GSSIGQATYY119 pKa = 9.76TITEE123 pKa = 4.44ACPCEE128 pKa = 4.04ATRR131 pKa = 11.84RR132 pKa = 11.84PGHH135 pKa = 6.43IPDD138 pKa = 4.42GFTTTVTSCEE148 pKa = 4.31VCHH151 pKa = 6.62HH152 pKa = 6.95GGPTTMTMTVPCTTGPYY169 pKa = 9.24ATVTPHH175 pKa = 6.9IGPTGAAGGWGDD187 pKa = 3.5WDD189 pKa = 4.25GSGSDD194 pKa = 3.99PAEE197 pKa = 4.18SGAWGDD203 pKa = 3.53WDD205 pKa = 4.69GSGPDD210 pKa = 3.93PAKK213 pKa = 10.69SGAWGDD219 pKa = 3.78WNSADD224 pKa = 4.38PDD226 pKa = 4.23PAKK229 pKa = 10.75SGAWGDD235 pKa = 3.57WSSGADD241 pKa = 3.61PTKK244 pKa = 10.75SGAWGDD250 pKa = 3.85WNSADD255 pKa = 4.38PDD257 pKa = 4.23PAKK260 pKa = 10.75SGAWGDD266 pKa = 3.57WSSGADD272 pKa = 3.55PTATGAWGDD281 pKa = 3.78WKK283 pKa = 11.2SGDD286 pKa = 4.17PEE288 pKa = 4.08PTATGTWADD297 pKa = 3.64WEE299 pKa = 4.63GSDD302 pKa = 5.32PEE304 pKa = 4.26PTEE307 pKa = 4.31GGSWDD312 pKa = 4.0DD313 pKa = 3.15WDD315 pKa = 4.13KK316 pKa = 11.7SGSGGSGGSGSGSGGSGAGGASADD340 pKa = 3.27KK341 pKa = 9.91WDD343 pKa = 4.03NGSAGGGSPASPSVTPVSPGGVTPAQYY370 pKa = 10.3KK371 pKa = 10.47GSADD375 pKa = 3.2KK376 pKa = 11.45VGFAVSGVLAVAVGCLAFMLL396 pKa = 4.75
MM1 pKa = 7.16KK2 pKa = 10.19RR3 pKa = 11.84STRR6 pKa = 11.84AMPVAALLMAAANAQTTIINDD27 pKa = 3.87PADD30 pKa = 4.33PDD32 pKa = 3.65ATTVYY37 pKa = 10.99YY38 pKa = 7.95STCPTALTTTTTLSSTITFTCPGGEE63 pKa = 4.06CSGPTITPPPHH74 pKa = 6.53GSAQVSEE81 pKa = 4.21ILAFTTTLPDD91 pKa = 3.41GSVQEE96 pKa = 3.75IHH98 pKa = 6.95EE99 pKa = 4.96YY100 pKa = 8.35ITMYY104 pKa = 9.76PAIVSDD110 pKa = 4.46GSSIGQATYY119 pKa = 9.76TITEE123 pKa = 4.44ACPCEE128 pKa = 4.04ATRR131 pKa = 11.84RR132 pKa = 11.84PGHH135 pKa = 6.43IPDD138 pKa = 4.42GFTTTVTSCEE148 pKa = 4.31VCHH151 pKa = 6.62HH152 pKa = 6.95GGPTTMTMTVPCTTGPYY169 pKa = 9.24ATVTPHH175 pKa = 6.9IGPTGAAGGWGDD187 pKa = 3.5WDD189 pKa = 4.25GSGSDD194 pKa = 3.99PAEE197 pKa = 4.18SGAWGDD203 pKa = 3.53WDD205 pKa = 4.69GSGPDD210 pKa = 3.93PAKK213 pKa = 10.69SGAWGDD219 pKa = 3.78WNSADD224 pKa = 4.38PDD226 pKa = 4.23PAKK229 pKa = 10.75SGAWGDD235 pKa = 3.57WSSGADD241 pKa = 3.61PTKK244 pKa = 10.75SGAWGDD250 pKa = 3.85WNSADD255 pKa = 4.38PDD257 pKa = 4.23PAKK260 pKa = 10.75SGAWGDD266 pKa = 3.57WSSGADD272 pKa = 3.55PTATGAWGDD281 pKa = 3.78WKK283 pKa = 11.2SGDD286 pKa = 4.17PEE288 pKa = 4.08PTATGTWADD297 pKa = 3.64WEE299 pKa = 4.63GSDD302 pKa = 5.32PEE304 pKa = 4.26PTEE307 pKa = 4.31GGSWDD312 pKa = 4.0DD313 pKa = 3.15WDD315 pKa = 4.13KK316 pKa = 11.7SGSGGSGGSGSGSGGSGAGGASADD340 pKa = 3.27KK341 pKa = 9.91WDD343 pKa = 4.03NGSAGGGSPASPSVTPVSPGGVTPAQYY370 pKa = 10.3KK371 pKa = 10.47GSADD375 pKa = 3.2KK376 pKa = 11.45VGFAVSGVLAVAVGCLAFMLL396 pKa = 4.75
Molecular weight: 39.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D2F4Q3|A0A0D2F4Q3_9EURO Uncharacterized protein OS=Phialophora americana OX=5601 GN=PV04_09760 PE=4 SV=1
MM1 pKa = 7.16ATLARR6 pKa = 11.84VFQEE10 pKa = 3.78ACRR13 pKa = 11.84SRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84DD18 pKa = 3.46RR19 pKa = 11.84VQHH22 pKa = 6.49ACACLPSLTRR32 pKa = 11.84SGEE35 pKa = 3.85LTATLKK41 pKa = 10.73RR42 pKa = 11.84RR43 pKa = 11.84GPPSTSFHH51 pKa = 6.25QISCRR56 pKa = 11.84SQPPGRR62 pKa = 11.84RR63 pKa = 11.84VKK65 pKa = 10.6SRR67 pKa = 11.84SASTSSGPSRR77 pKa = 11.84RR78 pKa = 11.84AVTVTSDD85 pKa = 3.18DD86 pKa = 3.3GRR88 pKa = 11.84YY89 pKa = 8.98NWSEE93 pKa = 3.74LSTGEE98 pKa = 3.67KK99 pKa = 9.89AARR102 pKa = 11.84GTQQTFNFLLVSLGLVGTITVAYY125 pKa = 9.56FLYY128 pKa = 10.59QEE130 pKa = 4.67LLAPSSSTIQFNAAVSRR147 pKa = 11.84IKK149 pKa = 10.81SSSEE153 pKa = 3.25CRR155 pKa = 11.84ALLGPSSQILAYY167 pKa = 10.32GEE169 pKa = 4.11PTSSKK174 pKa = 8.45WARR177 pKa = 11.84ARR179 pKa = 11.84PLAHH183 pKa = 5.68STEE186 pKa = 3.92VDD188 pKa = 3.46RR189 pKa = 11.84NGTTHH194 pKa = 7.0LRR196 pKa = 11.84MHH198 pKa = 6.77FNVEE202 pKa = 4.08GRR204 pKa = 11.84DD205 pKa = 3.59SGVKK209 pKa = 10.18GVVNVHH215 pKa = 4.62MTRR218 pKa = 11.84PKK220 pKa = 10.84DD221 pKa = 3.61GDD223 pKa = 3.47RR224 pKa = 11.84LEE226 pKa = 4.16YY227 pKa = 10.4QLLSLSVKK235 pKa = 9.71GHH237 pKa = 4.41EE238 pKa = 4.28TVYY241 pKa = 11.01LEE243 pKa = 4.04NKK245 pKa = 8.84AAEE248 pKa = 4.14RR249 pKa = 11.84GIKK252 pKa = 9.46GQAAKK257 pKa = 10.04MFGVQWRR264 pKa = 3.68
MM1 pKa = 7.16ATLARR6 pKa = 11.84VFQEE10 pKa = 3.78ACRR13 pKa = 11.84SRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84DD18 pKa = 3.46RR19 pKa = 11.84VQHH22 pKa = 6.49ACACLPSLTRR32 pKa = 11.84SGEE35 pKa = 3.85LTATLKK41 pKa = 10.73RR42 pKa = 11.84RR43 pKa = 11.84GPPSTSFHH51 pKa = 6.25QISCRR56 pKa = 11.84SQPPGRR62 pKa = 11.84RR63 pKa = 11.84VKK65 pKa = 10.6SRR67 pKa = 11.84SASTSSGPSRR77 pKa = 11.84RR78 pKa = 11.84AVTVTSDD85 pKa = 3.18DD86 pKa = 3.3GRR88 pKa = 11.84YY89 pKa = 8.98NWSEE93 pKa = 3.74LSTGEE98 pKa = 3.67KK99 pKa = 9.89AARR102 pKa = 11.84GTQQTFNFLLVSLGLVGTITVAYY125 pKa = 9.56FLYY128 pKa = 10.59QEE130 pKa = 4.67LLAPSSSTIQFNAAVSRR147 pKa = 11.84IKK149 pKa = 10.81SSSEE153 pKa = 3.25CRR155 pKa = 11.84ALLGPSSQILAYY167 pKa = 10.32GEE169 pKa = 4.11PTSSKK174 pKa = 8.45WARR177 pKa = 11.84ARR179 pKa = 11.84PLAHH183 pKa = 5.68STEE186 pKa = 3.92VDD188 pKa = 3.46RR189 pKa = 11.84NGTTHH194 pKa = 7.0LRR196 pKa = 11.84MHH198 pKa = 6.77FNVEE202 pKa = 4.08GRR204 pKa = 11.84DD205 pKa = 3.59SGVKK209 pKa = 10.18GVVNVHH215 pKa = 4.62MTRR218 pKa = 11.84PKK220 pKa = 10.84DD221 pKa = 3.61GDD223 pKa = 3.47RR224 pKa = 11.84LEE226 pKa = 4.16YY227 pKa = 10.4QLLSLSVKK235 pKa = 9.71GHH237 pKa = 4.41EE238 pKa = 4.28TVYY241 pKa = 11.01LEE243 pKa = 4.04NKK245 pKa = 8.84AAEE248 pKa = 4.14RR249 pKa = 11.84GIKK252 pKa = 9.46GQAAKK257 pKa = 10.04MFGVQWRR264 pKa = 3.68
Molecular weight: 29.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5818011 |
55 |
6403 |
505.8 |
55.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.804 ± 0.019 | 1.21 ± 0.008 |
5.731 ± 0.016 | 6.048 ± 0.019 |
3.675 ± 0.012 | 6.843 ± 0.02 |
2.475 ± 0.01 | 4.732 ± 0.015 |
4.662 ± 0.019 | 8.95 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.15 ± 0.007 | 3.505 ± 0.012 |
6.203 ± 0.024 | 4.165 ± 0.017 |
6.324 ± 0.02 | 8.161 ± 0.026 |
6.039 ± 0.016 | 6.159 ± 0.016 |
1.454 ± 0.008 | 2.709 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
