
Tortoise microvirus 11
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.07
Get precalculated fractions of proteins
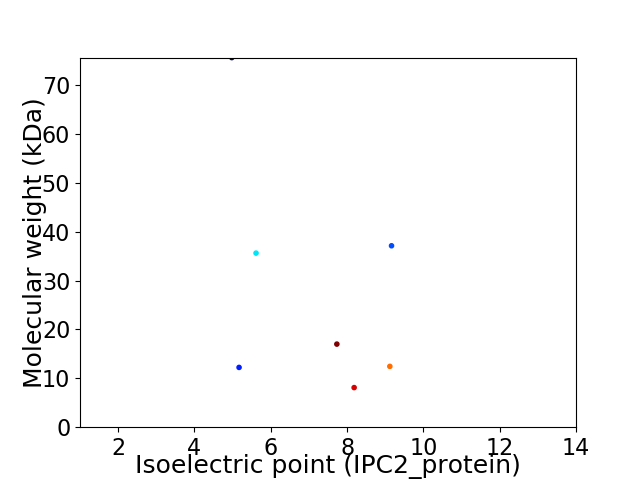
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVW5|A0A4V1FVW5_9VIRU Uncharacterized protein OS=Tortoise microvirus 11 OX=2583110 PE=4 SV=1
MM1 pKa = 7.43SNKK4 pKa = 8.61KK5 pKa = 8.08TFGGDD10 pKa = 3.09RR11 pKa = 11.84LGTGAGFTVSQRR23 pKa = 11.84AYY25 pKa = 9.53EE26 pKa = 4.19RR27 pKa = 11.84SSHH30 pKa = 7.03DD31 pKa = 3.29LGNVFRR37 pKa = 11.84SNLGVGMIMPCYY49 pKa = 10.03VDD51 pKa = 4.03FVKK54 pKa = 10.85RR55 pKa = 11.84GDD57 pKa = 3.36VWEE60 pKa = 4.4KK61 pKa = 11.08DD62 pKa = 3.09ITQHH66 pKa = 6.91ILTHH70 pKa = 5.85PTNGPIFGSYY80 pKa = 7.41EE81 pKa = 3.65CRR83 pKa = 11.84VAVFTGDD90 pKa = 2.87VRR92 pKa = 11.84LYY94 pKa = 11.06NKK96 pKa = 9.54QLHH99 pKa = 6.4NNTKK103 pKa = 10.85GIGLDD108 pKa = 3.3MKK110 pKa = 10.94SVIFPKK116 pKa = 9.82MRR118 pKa = 11.84LNGRR122 pKa = 11.84NIQWAEE128 pKa = 3.97NQDD131 pKa = 3.97LNAQQVNQTSLLATTGIRR149 pKa = 11.84GLGTLEE155 pKa = 3.97KK156 pKa = 10.33ADD158 pKa = 4.14GIGRR162 pKa = 11.84VEE164 pKa = 3.99IQRR167 pKa = 11.84NAMWVMMYY175 pKa = 10.21YY176 pKa = 10.4EE177 pKa = 4.84IYY179 pKa = 10.5SEE181 pKa = 4.25YY182 pKa = 10.24FANKK186 pKa = 8.74QEE188 pKa = 4.38EE189 pKa = 4.28IGVVIGGSEE198 pKa = 4.22VKK200 pKa = 9.62GTASVDD206 pKa = 3.94RR207 pKa = 11.84IQGRR211 pKa = 11.84SIFSGSNEE219 pKa = 3.51WTITQNNIIEE229 pKa = 4.38PNGNAGLQPQHH240 pKa = 6.27TYY242 pKa = 10.78RR243 pKa = 11.84ITGSGIGEE251 pKa = 3.76QTVAVKK257 pKa = 10.19IANNTYY263 pKa = 10.43VLEE266 pKa = 4.54KK267 pKa = 9.87FDD269 pKa = 4.26KK270 pKa = 11.1FIDD273 pKa = 4.15FKK275 pKa = 11.54YY276 pKa = 10.05SASGTWLEE284 pKa = 4.03FTVAQGGPQTITADD298 pKa = 3.22ANGRR302 pKa = 11.84LFQASGRR309 pKa = 11.84EE310 pKa = 3.94ITMGTDD316 pKa = 2.85IEE318 pKa = 4.54LLEE321 pKa = 4.77FPLSNIDD328 pKa = 3.96DD329 pKa = 3.7MRR331 pKa = 11.84EE332 pKa = 3.78KK333 pKa = 11.04VFAQPKK339 pKa = 8.09TSPLIIGYY347 pKa = 10.03YY348 pKa = 10.16GDD350 pKa = 4.03EE351 pKa = 4.27KK352 pKa = 11.09HH353 pKa = 6.31GLPYY357 pKa = 10.78DD358 pKa = 3.54QVTGQTSLDD367 pKa = 3.63STGKK371 pKa = 9.9PNTTSNMNSITDD383 pKa = 3.6WAGLGIATYY392 pKa = 10.52KK393 pKa = 9.95ADD395 pKa = 4.87RR396 pKa = 11.84WQTWLNSEE404 pKa = 4.17WIEE407 pKa = 4.03NVNTISSVDD416 pKa = 3.43TSSGSFSMDD425 pKa = 2.64ALAVAKK431 pKa = 10.24RR432 pKa = 11.84LWKK435 pKa = 10.52FNNDD439 pKa = 2.76VVAGGGSYY447 pKa = 10.51QDD449 pKa = 2.62WLMAVDD455 pKa = 4.51GMEE458 pKa = 4.21TYY460 pKa = 10.25GAPEE464 pKa = 3.8MPVYY468 pKa = 10.59RR469 pKa = 11.84GGCSSMITFDD479 pKa = 3.72EE480 pKa = 4.93VISSSEE486 pKa = 4.09YY487 pKa = 10.38GDD489 pKa = 3.67QPLGTLGGQGVMKK502 pKa = 10.07NVKK505 pKa = 9.25GGHH508 pKa = 3.88VRR510 pKa = 11.84FRR512 pKa = 11.84AEE514 pKa = 4.03EE515 pKa = 3.67NGMIMVVVWITPIIDD530 pKa = 3.54YY531 pKa = 10.3YY532 pKa = 11.14QGNKK536 pKa = 7.65WFTKK540 pKa = 10.52LEE542 pKa = 4.14TMDD545 pKa = 6.06DD546 pKa = 2.9IHH548 pKa = 8.74KK549 pKa = 10.15PIFDD553 pKa = 5.02AIGFQEE559 pKa = 4.12KK560 pKa = 7.91TTDD563 pKa = 4.97QMAAWDD569 pKa = 4.4TIVNEE574 pKa = 4.96DD575 pKa = 3.2GTEE578 pKa = 4.07TFFSAGKK585 pKa = 8.91EE586 pKa = 4.18PAWTEE591 pKa = 3.36YY592 pKa = 6.32WTRR595 pKa = 11.84NNEE598 pKa = 4.0NYY600 pKa = 10.84GSFTRR605 pKa = 11.84HH606 pKa = 4.54NDD608 pKa = 2.72EE609 pKa = 4.86RR610 pKa = 11.84YY611 pKa = 8.3MVLNRR616 pKa = 11.84DD617 pKa = 3.24YY618 pKa = 11.64TMNEE622 pKa = 3.27EE623 pKa = 4.32DD624 pKa = 5.59GRR626 pKa = 11.84IEE628 pKa = 5.71DD629 pKa = 3.87LTTYY633 pKa = 10.52IDD635 pKa = 3.69PTKK638 pKa = 10.43FLYY641 pKa = 10.04PFAYY645 pKa = 9.24TGLQYY650 pKa = 11.31GPFWVQIGFDD660 pKa = 3.79TITRR664 pKa = 11.84RR665 pKa = 11.84IMSGAKK671 pKa = 9.08MPTLL675 pKa = 3.75
MM1 pKa = 7.43SNKK4 pKa = 8.61KK5 pKa = 8.08TFGGDD10 pKa = 3.09RR11 pKa = 11.84LGTGAGFTVSQRR23 pKa = 11.84AYY25 pKa = 9.53EE26 pKa = 4.19RR27 pKa = 11.84SSHH30 pKa = 7.03DD31 pKa = 3.29LGNVFRR37 pKa = 11.84SNLGVGMIMPCYY49 pKa = 10.03VDD51 pKa = 4.03FVKK54 pKa = 10.85RR55 pKa = 11.84GDD57 pKa = 3.36VWEE60 pKa = 4.4KK61 pKa = 11.08DD62 pKa = 3.09ITQHH66 pKa = 6.91ILTHH70 pKa = 5.85PTNGPIFGSYY80 pKa = 7.41EE81 pKa = 3.65CRR83 pKa = 11.84VAVFTGDD90 pKa = 2.87VRR92 pKa = 11.84LYY94 pKa = 11.06NKK96 pKa = 9.54QLHH99 pKa = 6.4NNTKK103 pKa = 10.85GIGLDD108 pKa = 3.3MKK110 pKa = 10.94SVIFPKK116 pKa = 9.82MRR118 pKa = 11.84LNGRR122 pKa = 11.84NIQWAEE128 pKa = 3.97NQDD131 pKa = 3.97LNAQQVNQTSLLATTGIRR149 pKa = 11.84GLGTLEE155 pKa = 3.97KK156 pKa = 10.33ADD158 pKa = 4.14GIGRR162 pKa = 11.84VEE164 pKa = 3.99IQRR167 pKa = 11.84NAMWVMMYY175 pKa = 10.21YY176 pKa = 10.4EE177 pKa = 4.84IYY179 pKa = 10.5SEE181 pKa = 4.25YY182 pKa = 10.24FANKK186 pKa = 8.74QEE188 pKa = 4.38EE189 pKa = 4.28IGVVIGGSEE198 pKa = 4.22VKK200 pKa = 9.62GTASVDD206 pKa = 3.94RR207 pKa = 11.84IQGRR211 pKa = 11.84SIFSGSNEE219 pKa = 3.51WTITQNNIIEE229 pKa = 4.38PNGNAGLQPQHH240 pKa = 6.27TYY242 pKa = 10.78RR243 pKa = 11.84ITGSGIGEE251 pKa = 3.76QTVAVKK257 pKa = 10.19IANNTYY263 pKa = 10.43VLEE266 pKa = 4.54KK267 pKa = 9.87FDD269 pKa = 4.26KK270 pKa = 11.1FIDD273 pKa = 4.15FKK275 pKa = 11.54YY276 pKa = 10.05SASGTWLEE284 pKa = 4.03FTVAQGGPQTITADD298 pKa = 3.22ANGRR302 pKa = 11.84LFQASGRR309 pKa = 11.84EE310 pKa = 3.94ITMGTDD316 pKa = 2.85IEE318 pKa = 4.54LLEE321 pKa = 4.77FPLSNIDD328 pKa = 3.96DD329 pKa = 3.7MRR331 pKa = 11.84EE332 pKa = 3.78KK333 pKa = 11.04VFAQPKK339 pKa = 8.09TSPLIIGYY347 pKa = 10.03YY348 pKa = 10.16GDD350 pKa = 4.03EE351 pKa = 4.27KK352 pKa = 11.09HH353 pKa = 6.31GLPYY357 pKa = 10.78DD358 pKa = 3.54QVTGQTSLDD367 pKa = 3.63STGKK371 pKa = 9.9PNTTSNMNSITDD383 pKa = 3.6WAGLGIATYY392 pKa = 10.52KK393 pKa = 9.95ADD395 pKa = 4.87RR396 pKa = 11.84WQTWLNSEE404 pKa = 4.17WIEE407 pKa = 4.03NVNTISSVDD416 pKa = 3.43TSSGSFSMDD425 pKa = 2.64ALAVAKK431 pKa = 10.24RR432 pKa = 11.84LWKK435 pKa = 10.52FNNDD439 pKa = 2.76VVAGGGSYY447 pKa = 10.51QDD449 pKa = 2.62WLMAVDD455 pKa = 4.51GMEE458 pKa = 4.21TYY460 pKa = 10.25GAPEE464 pKa = 3.8MPVYY468 pKa = 10.59RR469 pKa = 11.84GGCSSMITFDD479 pKa = 3.72EE480 pKa = 4.93VISSSEE486 pKa = 4.09YY487 pKa = 10.38GDD489 pKa = 3.67QPLGTLGGQGVMKK502 pKa = 10.07NVKK505 pKa = 9.25GGHH508 pKa = 3.88VRR510 pKa = 11.84FRR512 pKa = 11.84AEE514 pKa = 4.03EE515 pKa = 3.67NGMIMVVVWITPIIDD530 pKa = 3.54YY531 pKa = 10.3YY532 pKa = 11.14QGNKK536 pKa = 7.65WFTKK540 pKa = 10.52LEE542 pKa = 4.14TMDD545 pKa = 6.06DD546 pKa = 2.9IHH548 pKa = 8.74KK549 pKa = 10.15PIFDD553 pKa = 5.02AIGFQEE559 pKa = 4.12KK560 pKa = 7.91TTDD563 pKa = 4.97QMAAWDD569 pKa = 4.4TIVNEE574 pKa = 4.96DD575 pKa = 3.2GTEE578 pKa = 4.07TFFSAGKK585 pKa = 8.91EE586 pKa = 4.18PAWTEE591 pKa = 3.36YY592 pKa = 6.32WTRR595 pKa = 11.84NNEE598 pKa = 4.0NYY600 pKa = 10.84GSFTRR605 pKa = 11.84HH606 pKa = 4.54NDD608 pKa = 2.72EE609 pKa = 4.86RR610 pKa = 11.84YY611 pKa = 8.3MVLNRR616 pKa = 11.84DD617 pKa = 3.24YY618 pKa = 11.64TMNEE622 pKa = 3.27EE623 pKa = 4.32DD624 pKa = 5.59GRR626 pKa = 11.84IEE628 pKa = 5.71DD629 pKa = 3.87LTTYY633 pKa = 10.52IDD635 pKa = 3.69PTKK638 pKa = 10.43FLYY641 pKa = 10.04PFAYY645 pKa = 9.24TGLQYY650 pKa = 11.31GPFWVQIGFDD660 pKa = 3.79TITRR664 pKa = 11.84RR665 pKa = 11.84IMSGAKK671 pKa = 9.08MPTLL675 pKa = 3.75
Molecular weight: 75.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W6J3|A0A4P8W6J3_9VIRU Uncharacterized protein OS=Tortoise microvirus 11 OX=2583110 PE=4 SV=1
MM1 pKa = 7.99RR2 pKa = 11.84NPGFGSRR9 pKa = 11.84KK10 pKa = 9.46RR11 pKa = 11.84LLKK14 pKa = 10.49INNSEE19 pKa = 4.14VGDD22 pKa = 4.02DD23 pKa = 4.63LMTKK27 pKa = 10.16LRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84TEE33 pKa = 4.05DD34 pKa = 3.19NKK36 pKa = 11.59DD37 pKa = 3.18MNTPGRR43 pKa = 11.84PLLYY47 pKa = 9.42TEE49 pKa = 5.13RR50 pKa = 11.84KK51 pKa = 9.32HH52 pKa = 7.06GVIPEE57 pKa = 4.19GNPRR61 pKa = 11.84TDD63 pKa = 2.62RR64 pKa = 11.84WDD66 pKa = 3.29IAQEE70 pKa = 3.8ATDD73 pKa = 3.78YY74 pKa = 10.77IARR77 pKa = 11.84SKK79 pKa = 10.37AARR82 pKa = 11.84RR83 pKa = 11.84DD84 pKa = 3.71AFDD87 pKa = 3.57KK88 pKa = 11.42AGGTEE93 pKa = 4.16GEE95 pKa = 4.12RR96 pKa = 11.84KK97 pKa = 9.38ILAEE101 pKa = 3.65RR102 pKa = 11.84ASKK105 pKa = 11.08NEE107 pKa = 3.78NTT109 pKa = 4.25
MM1 pKa = 7.99RR2 pKa = 11.84NPGFGSRR9 pKa = 11.84KK10 pKa = 9.46RR11 pKa = 11.84LLKK14 pKa = 10.49INNSEE19 pKa = 4.14VGDD22 pKa = 4.02DD23 pKa = 4.63LMTKK27 pKa = 10.16LRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84TEE33 pKa = 4.05DD34 pKa = 3.19NKK36 pKa = 11.59DD37 pKa = 3.18MNTPGRR43 pKa = 11.84PLLYY47 pKa = 9.42TEE49 pKa = 5.13RR50 pKa = 11.84KK51 pKa = 9.32HH52 pKa = 7.06GVIPEE57 pKa = 4.19GNPRR61 pKa = 11.84TDD63 pKa = 2.62RR64 pKa = 11.84WDD66 pKa = 3.29IAQEE70 pKa = 3.8ATDD73 pKa = 3.78YY74 pKa = 10.77IARR77 pKa = 11.84SKK79 pKa = 10.37AARR82 pKa = 11.84RR83 pKa = 11.84DD84 pKa = 3.71AFDD87 pKa = 3.57KK88 pKa = 11.42AGGTEE93 pKa = 4.16GEE95 pKa = 4.12RR96 pKa = 11.84KK97 pKa = 9.38ILAEE101 pKa = 3.65RR102 pKa = 11.84ASKK105 pKa = 11.08NEE107 pKa = 3.78NTT109 pKa = 4.25
Molecular weight: 12.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1752 |
70 |
675 |
250.3 |
28.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.336 ± 1.707 | 0.628 ± 0.19 |
4.338 ± 0.846 | 9.247 ± 1.356 |
3.025 ± 0.639 | 8.904 ± 0.905 |
1.427 ± 0.327 | 7.705 ± 0.439 |
8.847 ± 2.18 | 5.594 ± 0.475 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.253 ± 0.443 | 6.221 ± 0.282 |
2.74 ± 0.446 | 3.881 ± 0.621 |
5.422 ± 0.784 | 5.879 ± 0.68 |
6.906 ± 0.806 | 4.167 ± 0.835 |
1.769 ± 0.392 | 3.71 ± 0.599 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
