
Woodlouse hunter spider associated circular virus 1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.58
Get precalculated fractions of proteins
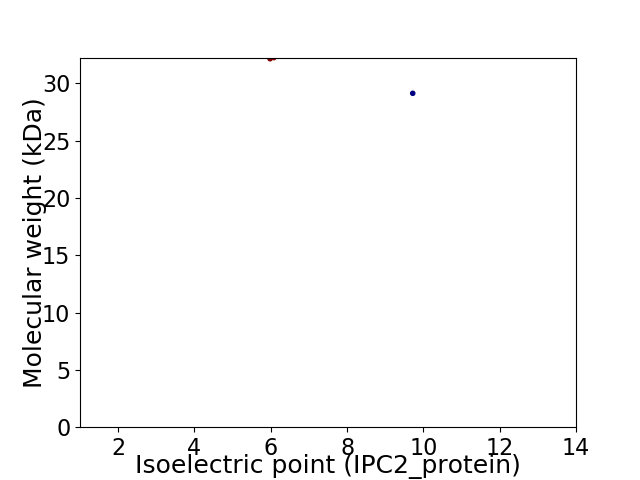
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BPE4|A0A346BPE4_9VIRU Putative capsid protein OS=Woodlouse hunter spider associated circular virus 1 OX=2293310 PE=4 SV=1
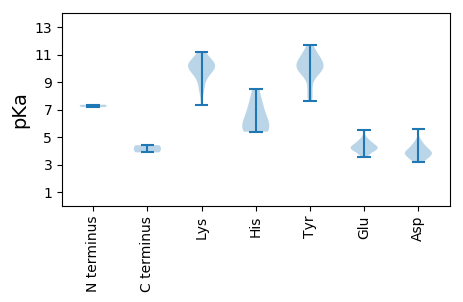
MM1 pKa = 7.18SRR3 pKa = 11.84SRR5 pKa = 11.84SYY7 pKa = 11.3CFTWNNYY14 pKa = 7.59PVNHH18 pKa = 6.27QDD20 pKa = 3.15VLDD23 pKa = 4.05RR24 pKa = 11.84LGYY27 pKa = 10.19RR28 pKa = 11.84YY29 pKa = 10.12VCYY32 pKa = 9.99GYY34 pKa = 10.5EE35 pKa = 3.88WAPTTRR41 pKa = 11.84TPHH44 pKa = 5.44LQGFIYY50 pKa = 10.29FRR52 pKa = 11.84DD53 pKa = 3.89AKK55 pKa = 9.65TMRR58 pKa = 11.84TVVRR62 pKa = 11.84GLPGVHH68 pKa = 6.29IEE70 pKa = 4.33SARR73 pKa = 11.84GTPAQCIEE81 pKa = 4.13YY82 pKa = 9.77CRR84 pKa = 11.84KK85 pKa = 10.26DD86 pKa = 3.29GEE88 pKa = 4.34FIEE91 pKa = 5.52FGEE94 pKa = 4.72APQTPQEE101 pKa = 3.98IGEE104 pKa = 4.28AEE106 pKa = 4.1KK107 pKa = 10.97DD108 pKa = 3.4RR109 pKa = 11.84YY110 pKa = 8.08LTAWDD115 pKa = 3.97LAKK118 pKa = 10.58AGQIDD123 pKa = 4.55EE124 pKa = 4.76IDD126 pKa = 3.63PEE128 pKa = 4.21LRR130 pKa = 11.84LRR132 pKa = 11.84HH133 pKa = 5.48YY134 pKa = 9.62STLKK138 pKa = 10.84SIAKK142 pKa = 9.74DD143 pKa = 3.54YY144 pKa = 9.92MVRR147 pKa = 11.84PPQLDD152 pKa = 3.84GVCGVWFWGPAGSGKK167 pKa = 7.33TTTANRR173 pKa = 11.84LYY175 pKa = 10.06PNAYY179 pKa = 10.13LKK181 pKa = 10.3PLNKK185 pKa = 9.14WWDD188 pKa = 3.87GYY190 pKa = 10.53QGEE193 pKa = 4.93DD194 pKa = 4.36VVICDD199 pKa = 4.68DD200 pKa = 3.25MSIFHH205 pKa = 7.21RR206 pKa = 11.84SLSDD210 pKa = 4.23LLKK213 pKa = 10.39HH214 pKa = 5.39WTDD217 pKa = 4.19FLPFIAEE224 pKa = 4.1VKK226 pKa = 10.3RR227 pKa = 11.84EE228 pKa = 4.07SVYY231 pKa = 10.3IRR233 pKa = 11.84PSKK236 pKa = 10.65FIVTSQYY243 pKa = 10.95SIEE246 pKa = 4.6EE247 pKa = 3.53IWAGDD252 pKa = 3.9EE253 pKa = 4.14KK254 pKa = 11.18SLEE257 pKa = 3.8AMRR260 pKa = 11.84RR261 pKa = 11.84RR262 pKa = 11.84FTVVEE267 pKa = 4.41KK268 pKa = 11.08VAGQDD273 pKa = 3.64IIMLL277 pKa = 4.4
MM1 pKa = 7.18SRR3 pKa = 11.84SRR5 pKa = 11.84SYY7 pKa = 11.3CFTWNNYY14 pKa = 7.59PVNHH18 pKa = 6.27QDD20 pKa = 3.15VLDD23 pKa = 4.05RR24 pKa = 11.84LGYY27 pKa = 10.19RR28 pKa = 11.84YY29 pKa = 10.12VCYY32 pKa = 9.99GYY34 pKa = 10.5EE35 pKa = 3.88WAPTTRR41 pKa = 11.84TPHH44 pKa = 5.44LQGFIYY50 pKa = 10.29FRR52 pKa = 11.84DD53 pKa = 3.89AKK55 pKa = 9.65TMRR58 pKa = 11.84TVVRR62 pKa = 11.84GLPGVHH68 pKa = 6.29IEE70 pKa = 4.33SARR73 pKa = 11.84GTPAQCIEE81 pKa = 4.13YY82 pKa = 9.77CRR84 pKa = 11.84KK85 pKa = 10.26DD86 pKa = 3.29GEE88 pKa = 4.34FIEE91 pKa = 5.52FGEE94 pKa = 4.72APQTPQEE101 pKa = 3.98IGEE104 pKa = 4.28AEE106 pKa = 4.1KK107 pKa = 10.97DD108 pKa = 3.4RR109 pKa = 11.84YY110 pKa = 8.08LTAWDD115 pKa = 3.97LAKK118 pKa = 10.58AGQIDD123 pKa = 4.55EE124 pKa = 4.76IDD126 pKa = 3.63PEE128 pKa = 4.21LRR130 pKa = 11.84LRR132 pKa = 11.84HH133 pKa = 5.48YY134 pKa = 9.62STLKK138 pKa = 10.84SIAKK142 pKa = 9.74DD143 pKa = 3.54YY144 pKa = 9.92MVRR147 pKa = 11.84PPQLDD152 pKa = 3.84GVCGVWFWGPAGSGKK167 pKa = 7.33TTTANRR173 pKa = 11.84LYY175 pKa = 10.06PNAYY179 pKa = 10.13LKK181 pKa = 10.3PLNKK185 pKa = 9.14WWDD188 pKa = 3.87GYY190 pKa = 10.53QGEE193 pKa = 4.93DD194 pKa = 4.36VVICDD199 pKa = 4.68DD200 pKa = 3.25MSIFHH205 pKa = 7.21RR206 pKa = 11.84SLSDD210 pKa = 4.23LLKK213 pKa = 10.39HH214 pKa = 5.39WTDD217 pKa = 4.19FLPFIAEE224 pKa = 4.1VKK226 pKa = 10.3RR227 pKa = 11.84EE228 pKa = 4.07SVYY231 pKa = 10.3IRR233 pKa = 11.84PSKK236 pKa = 10.65FIVTSQYY243 pKa = 10.95SIEE246 pKa = 4.6EE247 pKa = 3.53IWAGDD252 pKa = 3.9EE253 pKa = 4.14KK254 pKa = 11.18SLEE257 pKa = 3.8AMRR260 pKa = 11.84RR261 pKa = 11.84RR262 pKa = 11.84FTVVEE267 pKa = 4.41KK268 pKa = 11.08VAGQDD273 pKa = 3.64IIMLL277 pKa = 4.4
Molecular weight: 32.15 kDa
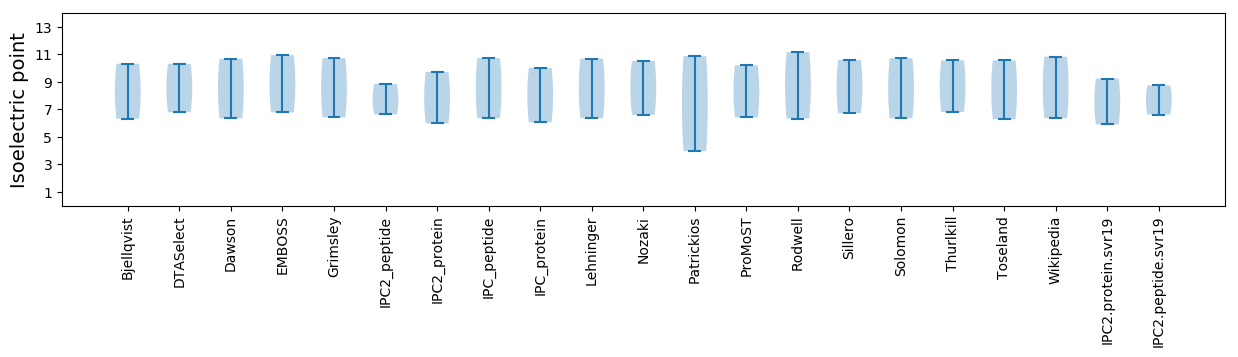
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BPE4|A0A346BPE4_9VIRU Putative capsid protein OS=Woodlouse hunter spider associated circular virus 1 OX=2293310 PE=4 SV=1
MM1 pKa = 7.35PRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84TAKK9 pKa = 8.67RR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 8.64RR13 pKa = 11.84RR14 pKa = 11.84TVKK17 pKa = 10.11RR18 pKa = 11.84RR19 pKa = 11.84SKK21 pKa = 10.21KK22 pKa = 9.66GRR24 pKa = 11.84RR25 pKa = 11.84LGKK28 pKa = 9.57RR29 pKa = 11.84QMDD32 pKa = 4.31IYY34 pKa = 10.12PSTGDD39 pKa = 3.43YY40 pKa = 11.04VYY42 pKa = 11.68GDD44 pKa = 3.73MHH46 pKa = 8.46RR47 pKa = 11.84MAKK50 pKa = 9.98KK51 pKa = 10.06IKK53 pKa = 8.41TSLGGSSSYY62 pKa = 9.63TASRR66 pKa = 11.84KK67 pKa = 10.07DD68 pKa = 3.34DD69 pKa = 4.43DD70 pKa = 3.92QWSMTQEE77 pKa = 4.21DD78 pKa = 4.79GIKK81 pKa = 10.48YY82 pKa = 8.23KK83 pKa = 10.32TINITYY89 pKa = 8.94KK90 pKa = 9.72KK91 pKa = 10.35SKK93 pKa = 9.84IGRR96 pKa = 11.84LTQRR100 pKa = 11.84LTSPGNLYY108 pKa = 10.69DD109 pKa = 3.75YY110 pKa = 9.68ATGGGASTQGRR121 pKa = 11.84QAVTDD126 pKa = 4.21ASAFNGGDD134 pKa = 3.62FNTLFQGLNNGVAITAIKK152 pKa = 10.41ASNQLFVKK160 pKa = 9.47QVKK163 pKa = 10.29HH164 pKa = 5.73EE165 pKa = 4.18MEE167 pKa = 4.55FNNAGPTTLEE177 pKa = 3.57MDD179 pKa = 3.94VYY181 pKa = 11.16ILIDD185 pKa = 3.89KK186 pKa = 8.1NTGGAISNPLTIWDD200 pKa = 3.75AALNSEE206 pKa = 4.56TNLVDD211 pKa = 5.55AIVEE215 pKa = 4.28AKK217 pKa = 10.03NKK219 pKa = 9.32PWLKK223 pKa = 9.2PTTLKK228 pKa = 11.04LFWFYY233 pKa = 10.69PKK235 pKa = 10.33SIPKK239 pKa = 9.96SIQTSRR245 pKa = 11.84IRR247 pKa = 11.84DD248 pKa = 3.76SNSTSHH254 pKa = 7.33TIPPRR259 pKa = 3.89
MM1 pKa = 7.35PRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84TAKK9 pKa = 8.67RR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 8.64RR13 pKa = 11.84RR14 pKa = 11.84TVKK17 pKa = 10.11RR18 pKa = 11.84RR19 pKa = 11.84SKK21 pKa = 10.21KK22 pKa = 9.66GRR24 pKa = 11.84RR25 pKa = 11.84LGKK28 pKa = 9.57RR29 pKa = 11.84QMDD32 pKa = 4.31IYY34 pKa = 10.12PSTGDD39 pKa = 3.43YY40 pKa = 11.04VYY42 pKa = 11.68GDD44 pKa = 3.73MHH46 pKa = 8.46RR47 pKa = 11.84MAKK50 pKa = 9.98KK51 pKa = 10.06IKK53 pKa = 8.41TSLGGSSSYY62 pKa = 9.63TASRR66 pKa = 11.84KK67 pKa = 10.07DD68 pKa = 3.34DD69 pKa = 4.43DD70 pKa = 3.92QWSMTQEE77 pKa = 4.21DD78 pKa = 4.79GIKK81 pKa = 10.48YY82 pKa = 8.23KK83 pKa = 10.32TINITYY89 pKa = 8.94KK90 pKa = 9.72KK91 pKa = 10.35SKK93 pKa = 9.84IGRR96 pKa = 11.84LTQRR100 pKa = 11.84LTSPGNLYY108 pKa = 10.69DD109 pKa = 3.75YY110 pKa = 9.68ATGGGASTQGRR121 pKa = 11.84QAVTDD126 pKa = 4.21ASAFNGGDD134 pKa = 3.62FNTLFQGLNNGVAITAIKK152 pKa = 10.41ASNQLFVKK160 pKa = 9.47QVKK163 pKa = 10.29HH164 pKa = 5.73EE165 pKa = 4.18MEE167 pKa = 4.55FNNAGPTTLEE177 pKa = 3.57MDD179 pKa = 3.94VYY181 pKa = 11.16ILIDD185 pKa = 3.89KK186 pKa = 8.1NTGGAISNPLTIWDD200 pKa = 3.75AALNSEE206 pKa = 4.56TNLVDD211 pKa = 5.55AIVEE215 pKa = 4.28AKK217 pKa = 10.03NKK219 pKa = 9.32PWLKK223 pKa = 9.2PTTLKK228 pKa = 11.04LFWFYY233 pKa = 10.69PKK235 pKa = 10.33SIPKK239 pKa = 9.96SIQTSRR245 pKa = 11.84IRR247 pKa = 11.84DD248 pKa = 3.76SNSTSHH254 pKa = 7.33TIPPRR259 pKa = 3.89
Molecular weight: 29.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
536 |
259 |
277 |
268.0 |
30.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.343 ± 0.149 | 1.119 ± 0.758 |
6.157 ± 0.247 | 4.664 ± 1.59 |
3.358 ± 0.444 | 7.276 ± 0.302 |
1.679 ± 0.353 | 6.343 ± 0.149 |
7.276 ± 1.609 | 6.53 ± 0.239 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.425 ± 0.188 | 3.918 ± 1.269 |
4.851 ± 0.409 | 3.731 ± 0.088 |
7.836 ± 0.184 | 6.716 ± 0.942 |
7.649 ± 1.356 | 4.851 ± 0.932 |
2.425 ± 0.597 | 4.851 ± 0.67 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
