
Salarias fasciatus (Jewelled blenny) (Blennius fasciatus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha; Neoteleostei; Eurypterygia;
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
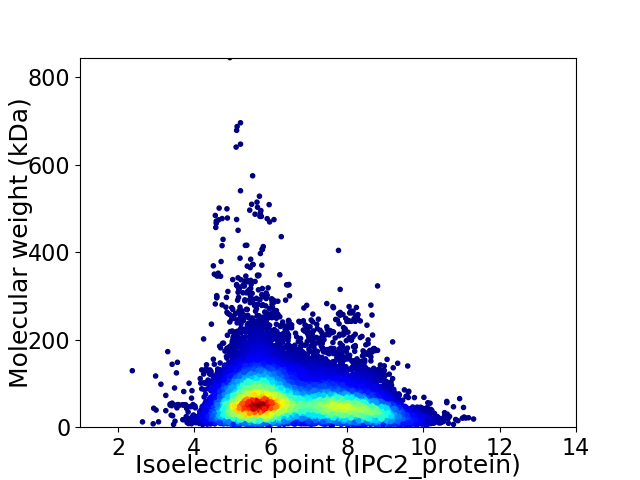
Virtual 2D-PAGE plot for 55738 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A672HYA4|A0A672HYA4_SALFA ANK_REP_REGION domain-containing protein OS=Salarias fasciatus OX=181472 GN=ankrd39 PE=4 SV=1
LL1 pKa = 8.29SFDD4 pKa = 3.53VTDD7 pKa = 4.17GKK9 pKa = 10.91DD10 pKa = 3.03RR11 pKa = 11.84IIFVTKK17 pKa = 9.82EE18 pKa = 3.87DD19 pKa = 3.6QEE21 pKa = 4.5APSNAEE27 pKa = 4.22LVADD31 pKa = 5.11DD32 pKa = 5.12PDD34 pKa = 4.84DD35 pKa = 4.49PYY37 pKa = 11.6EE38 pKa = 4.16EE39 pKa = 4.3QGLILPSGDD48 pKa = 3.72MNWNCPCLGGMASGPCGSQFKK69 pKa = 9.21EE70 pKa = 3.99ACSCFHH76 pKa = 6.25YY77 pKa = 10.82SKK79 pKa = 11.27EE80 pKa = 4.09EE81 pKa = 4.02VKK83 pKa = 10.98GSDD86 pKa = 3.11CLEE89 pKa = 3.99NVRR92 pKa = 11.84NLQEE96 pKa = 4.57CMQKK100 pKa = 10.11NPEE103 pKa = 4.71LYY105 pKa = 9.59PQEE108 pKa = 4.08EE109 pKa = 4.66DD110 pKa = 3.51EE111 pKa = 6.13DD112 pKa = 4.2VATQQEE118 pKa = 4.56SDD120 pKa = 3.6SALTSAAPPDD130 pKa = 3.93TSALSPEE137 pKa = 4.8SEE139 pKa = 4.28PSPAPSAVTPDD150 pKa = 3.4SSSATDD156 pKa = 3.64GQTASS161 pKa = 3.59
LL1 pKa = 8.29SFDD4 pKa = 3.53VTDD7 pKa = 4.17GKK9 pKa = 10.91DD10 pKa = 3.03RR11 pKa = 11.84IIFVTKK17 pKa = 9.82EE18 pKa = 3.87DD19 pKa = 3.6QEE21 pKa = 4.5APSNAEE27 pKa = 4.22LVADD31 pKa = 5.11DD32 pKa = 5.12PDD34 pKa = 4.84DD35 pKa = 4.49PYY37 pKa = 11.6EE38 pKa = 4.16EE39 pKa = 4.3QGLILPSGDD48 pKa = 3.72MNWNCPCLGGMASGPCGSQFKK69 pKa = 9.21EE70 pKa = 3.99ACSCFHH76 pKa = 6.25YY77 pKa = 10.82SKK79 pKa = 11.27EE80 pKa = 4.09EE81 pKa = 4.02VKK83 pKa = 10.98GSDD86 pKa = 3.11CLEE89 pKa = 3.99NVRR92 pKa = 11.84NLQEE96 pKa = 4.57CMQKK100 pKa = 10.11NPEE103 pKa = 4.71LYY105 pKa = 9.59PQEE108 pKa = 4.08EE109 pKa = 4.66DD110 pKa = 3.51EE111 pKa = 6.13DD112 pKa = 4.2VATQQEE118 pKa = 4.56SDD120 pKa = 3.6SALTSAAPPDD130 pKa = 3.93TSALSPEE137 pKa = 4.8SEE139 pKa = 4.28PSPAPSAVTPDD150 pKa = 3.4SSSATDD156 pKa = 3.64GQTASS161 pKa = 3.59
Molecular weight: 17.17 kDa
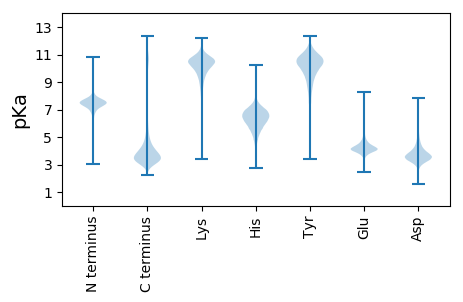
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A672J143|A0A672J143_SALFA Discs large (Drosophila) homolog-associated protein 2b OS=Salarias fasciatus OX=181472 PE=3 SV=1
SS1 pKa = 7.09SYY3 pKa = 11.57NEE5 pKa = 3.59VRR7 pKa = 11.84GQEE10 pKa = 3.89QHH12 pKa = 7.23LSEE15 pKa = 4.27QKK17 pKa = 10.24NRR19 pKa = 11.84EE20 pKa = 3.99YY21 pKa = 11.05SPKK24 pKa = 9.58TRR26 pKa = 11.84LKK28 pKa = 9.52TRR30 pKa = 11.84LKK32 pKa = 9.71TRR34 pKa = 11.84LKK36 pKa = 9.71TRR38 pKa = 11.84LKK40 pKa = 8.97TRR42 pKa = 11.84LRR44 pKa = 11.84TRR46 pKa = 11.84LKK48 pKa = 9.66TRR50 pKa = 11.84LKK52 pKa = 9.71TRR54 pKa = 11.84LKK56 pKa = 8.97TRR58 pKa = 11.84LRR60 pKa = 11.84TRR62 pKa = 11.84LKK64 pKa = 9.57TRR66 pKa = 11.84LKK68 pKa = 10.74NRR70 pKa = 11.84LKK72 pKa = 10.09TRR74 pKa = 11.84LKK76 pKa = 9.65TRR78 pKa = 11.84LKK80 pKa = 9.62TRR82 pKa = 11.84LKK84 pKa = 10.74NRR86 pKa = 11.84LKK88 pKa = 10.09TRR90 pKa = 11.84LKK92 pKa = 9.65TRR94 pKa = 11.84LKK96 pKa = 9.71TRR98 pKa = 11.84LKK100 pKa = 9.62TRR102 pKa = 11.84LKK104 pKa = 10.74NRR106 pKa = 11.84LKK108 pKa = 10.09TRR110 pKa = 11.84LKK112 pKa = 9.65TRR114 pKa = 11.84LKK116 pKa = 9.71TRR118 pKa = 11.84LKK120 pKa = 9.71TRR122 pKa = 11.84LKK124 pKa = 9.71TRR126 pKa = 11.84LKK128 pKa = 9.62TRR130 pKa = 11.84LKK132 pKa = 10.68NRR134 pKa = 11.84LKK136 pKa = 10.92NRR138 pKa = 11.84LKK140 pKa = 10.06TRR142 pKa = 11.84LKK144 pKa = 9.65TRR146 pKa = 11.84LKK148 pKa = 9.71TRR150 pKa = 11.84LKK152 pKa = 9.62TRR154 pKa = 11.84LKK156 pKa = 10.74NRR158 pKa = 11.84LKK160 pKa = 9.73TRR162 pKa = 11.84LRR164 pKa = 11.84TRR166 pKa = 11.84LKK168 pKa = 9.57TRR170 pKa = 11.84LKK172 pKa = 10.74NRR174 pKa = 11.84LKK176 pKa = 9.73TRR178 pKa = 11.84LRR180 pKa = 11.84TRR182 pKa = 11.84LKK184 pKa = 9.66TRR186 pKa = 11.84LKK188 pKa = 9.71TRR190 pKa = 11.84LKK192 pKa = 9.71TRR194 pKa = 11.84LKK196 pKa = 9.62TRR198 pKa = 11.84LKK200 pKa = 10.74NRR202 pKa = 11.84LKK204 pKa = 9.73TRR206 pKa = 11.84LRR208 pKa = 11.84TRR210 pKa = 11.84LKK212 pKa = 9.57TRR214 pKa = 11.84LKK216 pKa = 10.68NRR218 pKa = 11.84LKK220 pKa = 10.92NRR222 pKa = 11.84LKK224 pKa = 9.6TRR226 pKa = 11.84LRR228 pKa = 11.84TRR230 pKa = 11.84LKK232 pKa = 9.57TRR234 pKa = 11.84LKK236 pKa = 10.74NRR238 pKa = 11.84LKK240 pKa = 9.73TRR242 pKa = 11.84LRR244 pKa = 11.84TRR246 pKa = 11.84LKK248 pKa = 9.66TRR250 pKa = 11.84LKK252 pKa = 9.71TRR254 pKa = 11.84LKK256 pKa = 9.62TRR258 pKa = 11.84LKK260 pKa = 10.74NRR262 pKa = 11.84LKK264 pKa = 9.73TRR266 pKa = 11.84LRR268 pKa = 11.84TRR270 pKa = 11.84LKK272 pKa = 9.57TRR274 pKa = 11.84LKK276 pKa = 10.74NRR278 pKa = 11.84LKK280 pKa = 10.09TRR282 pKa = 11.84LKK284 pKa = 9.65TRR286 pKa = 11.84LKK288 pKa = 9.71TRR290 pKa = 11.84LKK292 pKa = 9.71TRR294 pKa = 11.84LKK296 pKa = 9.71TRR298 pKa = 11.84LKK300 pKa = 9.71TRR302 pKa = 11.84LKK304 pKa = 9.71TRR306 pKa = 11.84LKK308 pKa = 9.71TRR310 pKa = 11.84LKK312 pKa = 9.71TRR314 pKa = 11.84LKK316 pKa = 9.95TRR318 pKa = 11.84LKK320 pKa = 9.62TKK322 pKa = 10.58LKK324 pKa = 9.52TRR326 pKa = 11.84LKK328 pKa = 9.52TRR330 pKa = 11.84LKK332 pKa = 9.71TRR334 pKa = 11.84LKK336 pKa = 9.71TRR338 pKa = 11.84LKK340 pKa = 9.71TRR342 pKa = 11.84LKK344 pKa = 9.71TRR346 pKa = 11.84LKK348 pKa = 9.71TRR350 pKa = 11.84LKK352 pKa = 9.62TRR354 pKa = 11.84LKK356 pKa = 10.74NRR358 pKa = 11.84LKK360 pKa = 9.73TRR362 pKa = 11.84LRR364 pKa = 11.84TRR366 pKa = 11.84LKK368 pKa = 9.57TRR370 pKa = 11.84LKK372 pKa = 10.74NRR374 pKa = 11.84LKK376 pKa = 9.73TRR378 pKa = 11.84LRR380 pKa = 11.84TRR382 pKa = 11.84LKK384 pKa = 9.57TRR386 pKa = 11.84LKK388 pKa = 10.74NRR390 pKa = 11.84LKK392 pKa = 10.09TRR394 pKa = 11.84LKK396 pKa = 9.55TRR398 pKa = 11.84LKK400 pKa = 10.74NRR402 pKa = 11.84LKK404 pKa = 9.73TRR406 pKa = 11.84LRR408 pKa = 11.84TRR410 pKa = 11.84LKK412 pKa = 9.57TRR414 pKa = 11.84LKK416 pKa = 10.74NRR418 pKa = 11.84LKK420 pKa = 9.73TRR422 pKa = 11.84LRR424 pKa = 11.84TRR426 pKa = 11.84LKK428 pKa = 9.57TRR430 pKa = 11.84LKK432 pKa = 10.74NRR434 pKa = 11.84LKK436 pKa = 9.7TRR438 pKa = 11.84LRR440 pKa = 11.84TRR442 pKa = 11.84IKK444 pKa = 9.74TRR446 pKa = 11.84LKK448 pKa = 9.5TRR450 pKa = 11.84LKK452 pKa = 10.74NRR454 pKa = 11.84LKK456 pKa = 10.09TRR458 pKa = 11.84LKK460 pKa = 9.65TRR462 pKa = 11.84LKK464 pKa = 9.71TRR466 pKa = 11.84LKK468 pKa = 9.71TRR470 pKa = 11.84LKK472 pKa = 9.62TRR474 pKa = 11.84LKK476 pKa = 10.74NRR478 pKa = 11.84LKK480 pKa = 10.09TRR482 pKa = 11.84LKK484 pKa = 9.65TRR486 pKa = 11.84LKK488 pKa = 9.28TRR490 pKa = 11.84LRR492 pKa = 11.84TTLKK496 pKa = 9.59TRR498 pKa = 11.84LKK500 pKa = 10.46NRR502 pKa = 11.84LKK504 pKa = 9.62TRR506 pKa = 11.84VRR508 pKa = 11.84TRR510 pKa = 11.84LKK512 pKa = 9.79TRR514 pKa = 11.84LKK516 pKa = 9.71TRR518 pKa = 11.84LKK520 pKa = 9.71TRR522 pKa = 11.84LKK524 pKa = 9.71TRR526 pKa = 11.84LKK528 pKa = 8.97TRR530 pKa = 11.84LRR532 pKa = 11.84TRR534 pKa = 11.84LKK536 pKa = 9.57TRR538 pKa = 11.84LKK540 pKa = 10.74NRR542 pKa = 11.84LKK544 pKa = 10.09TRR546 pKa = 11.84LKK548 pKa = 9.65TRR550 pKa = 11.84LKK552 pKa = 9.71TRR554 pKa = 11.84LKK556 pKa = 8.97TRR558 pKa = 11.84LRR560 pKa = 11.84TRR562 pKa = 11.84LKK564 pKa = 10.71NRR566 pKa = 11.84LKK568 pKa = 10.09TRR570 pKa = 11.84LKK572 pKa = 9.65TRR574 pKa = 11.84LKK576 pKa = 9.38TRR578 pKa = 11.84LKK580 pKa = 10.87ARR582 pKa = 11.84LKK584 pKa = 9.77VAFISPHH591 pKa = 5.36VEE593 pKa = 3.7EE594 pKa = 4.6TASSRR599 pKa = 11.84RR600 pKa = 11.84KK601 pKa = 10.39EE602 pKa = 3.69EE603 pKa = 4.01DD604 pKa = 2.77
SS1 pKa = 7.09SYY3 pKa = 11.57NEE5 pKa = 3.59VRR7 pKa = 11.84GQEE10 pKa = 3.89QHH12 pKa = 7.23LSEE15 pKa = 4.27QKK17 pKa = 10.24NRR19 pKa = 11.84EE20 pKa = 3.99YY21 pKa = 11.05SPKK24 pKa = 9.58TRR26 pKa = 11.84LKK28 pKa = 9.52TRR30 pKa = 11.84LKK32 pKa = 9.71TRR34 pKa = 11.84LKK36 pKa = 9.71TRR38 pKa = 11.84LKK40 pKa = 8.97TRR42 pKa = 11.84LRR44 pKa = 11.84TRR46 pKa = 11.84LKK48 pKa = 9.66TRR50 pKa = 11.84LKK52 pKa = 9.71TRR54 pKa = 11.84LKK56 pKa = 8.97TRR58 pKa = 11.84LRR60 pKa = 11.84TRR62 pKa = 11.84LKK64 pKa = 9.57TRR66 pKa = 11.84LKK68 pKa = 10.74NRR70 pKa = 11.84LKK72 pKa = 10.09TRR74 pKa = 11.84LKK76 pKa = 9.65TRR78 pKa = 11.84LKK80 pKa = 9.62TRR82 pKa = 11.84LKK84 pKa = 10.74NRR86 pKa = 11.84LKK88 pKa = 10.09TRR90 pKa = 11.84LKK92 pKa = 9.65TRR94 pKa = 11.84LKK96 pKa = 9.71TRR98 pKa = 11.84LKK100 pKa = 9.62TRR102 pKa = 11.84LKK104 pKa = 10.74NRR106 pKa = 11.84LKK108 pKa = 10.09TRR110 pKa = 11.84LKK112 pKa = 9.65TRR114 pKa = 11.84LKK116 pKa = 9.71TRR118 pKa = 11.84LKK120 pKa = 9.71TRR122 pKa = 11.84LKK124 pKa = 9.71TRR126 pKa = 11.84LKK128 pKa = 9.62TRR130 pKa = 11.84LKK132 pKa = 10.68NRR134 pKa = 11.84LKK136 pKa = 10.92NRR138 pKa = 11.84LKK140 pKa = 10.06TRR142 pKa = 11.84LKK144 pKa = 9.65TRR146 pKa = 11.84LKK148 pKa = 9.71TRR150 pKa = 11.84LKK152 pKa = 9.62TRR154 pKa = 11.84LKK156 pKa = 10.74NRR158 pKa = 11.84LKK160 pKa = 9.73TRR162 pKa = 11.84LRR164 pKa = 11.84TRR166 pKa = 11.84LKK168 pKa = 9.57TRR170 pKa = 11.84LKK172 pKa = 10.74NRR174 pKa = 11.84LKK176 pKa = 9.73TRR178 pKa = 11.84LRR180 pKa = 11.84TRR182 pKa = 11.84LKK184 pKa = 9.66TRR186 pKa = 11.84LKK188 pKa = 9.71TRR190 pKa = 11.84LKK192 pKa = 9.71TRR194 pKa = 11.84LKK196 pKa = 9.62TRR198 pKa = 11.84LKK200 pKa = 10.74NRR202 pKa = 11.84LKK204 pKa = 9.73TRR206 pKa = 11.84LRR208 pKa = 11.84TRR210 pKa = 11.84LKK212 pKa = 9.57TRR214 pKa = 11.84LKK216 pKa = 10.68NRR218 pKa = 11.84LKK220 pKa = 10.92NRR222 pKa = 11.84LKK224 pKa = 9.6TRR226 pKa = 11.84LRR228 pKa = 11.84TRR230 pKa = 11.84LKK232 pKa = 9.57TRR234 pKa = 11.84LKK236 pKa = 10.74NRR238 pKa = 11.84LKK240 pKa = 9.73TRR242 pKa = 11.84LRR244 pKa = 11.84TRR246 pKa = 11.84LKK248 pKa = 9.66TRR250 pKa = 11.84LKK252 pKa = 9.71TRR254 pKa = 11.84LKK256 pKa = 9.62TRR258 pKa = 11.84LKK260 pKa = 10.74NRR262 pKa = 11.84LKK264 pKa = 9.73TRR266 pKa = 11.84LRR268 pKa = 11.84TRR270 pKa = 11.84LKK272 pKa = 9.57TRR274 pKa = 11.84LKK276 pKa = 10.74NRR278 pKa = 11.84LKK280 pKa = 10.09TRR282 pKa = 11.84LKK284 pKa = 9.65TRR286 pKa = 11.84LKK288 pKa = 9.71TRR290 pKa = 11.84LKK292 pKa = 9.71TRR294 pKa = 11.84LKK296 pKa = 9.71TRR298 pKa = 11.84LKK300 pKa = 9.71TRR302 pKa = 11.84LKK304 pKa = 9.71TRR306 pKa = 11.84LKK308 pKa = 9.71TRR310 pKa = 11.84LKK312 pKa = 9.71TRR314 pKa = 11.84LKK316 pKa = 9.95TRR318 pKa = 11.84LKK320 pKa = 9.62TKK322 pKa = 10.58LKK324 pKa = 9.52TRR326 pKa = 11.84LKK328 pKa = 9.52TRR330 pKa = 11.84LKK332 pKa = 9.71TRR334 pKa = 11.84LKK336 pKa = 9.71TRR338 pKa = 11.84LKK340 pKa = 9.71TRR342 pKa = 11.84LKK344 pKa = 9.71TRR346 pKa = 11.84LKK348 pKa = 9.71TRR350 pKa = 11.84LKK352 pKa = 9.62TRR354 pKa = 11.84LKK356 pKa = 10.74NRR358 pKa = 11.84LKK360 pKa = 9.73TRR362 pKa = 11.84LRR364 pKa = 11.84TRR366 pKa = 11.84LKK368 pKa = 9.57TRR370 pKa = 11.84LKK372 pKa = 10.74NRR374 pKa = 11.84LKK376 pKa = 9.73TRR378 pKa = 11.84LRR380 pKa = 11.84TRR382 pKa = 11.84LKK384 pKa = 9.57TRR386 pKa = 11.84LKK388 pKa = 10.74NRR390 pKa = 11.84LKK392 pKa = 10.09TRR394 pKa = 11.84LKK396 pKa = 9.55TRR398 pKa = 11.84LKK400 pKa = 10.74NRR402 pKa = 11.84LKK404 pKa = 9.73TRR406 pKa = 11.84LRR408 pKa = 11.84TRR410 pKa = 11.84LKK412 pKa = 9.57TRR414 pKa = 11.84LKK416 pKa = 10.74NRR418 pKa = 11.84LKK420 pKa = 9.73TRR422 pKa = 11.84LRR424 pKa = 11.84TRR426 pKa = 11.84LKK428 pKa = 9.57TRR430 pKa = 11.84LKK432 pKa = 10.74NRR434 pKa = 11.84LKK436 pKa = 9.7TRR438 pKa = 11.84LRR440 pKa = 11.84TRR442 pKa = 11.84IKK444 pKa = 9.74TRR446 pKa = 11.84LKK448 pKa = 9.5TRR450 pKa = 11.84LKK452 pKa = 10.74NRR454 pKa = 11.84LKK456 pKa = 10.09TRR458 pKa = 11.84LKK460 pKa = 9.65TRR462 pKa = 11.84LKK464 pKa = 9.71TRR466 pKa = 11.84LKK468 pKa = 9.71TRR470 pKa = 11.84LKK472 pKa = 9.62TRR474 pKa = 11.84LKK476 pKa = 10.74NRR478 pKa = 11.84LKK480 pKa = 10.09TRR482 pKa = 11.84LKK484 pKa = 9.65TRR486 pKa = 11.84LKK488 pKa = 9.28TRR490 pKa = 11.84LRR492 pKa = 11.84TTLKK496 pKa = 9.59TRR498 pKa = 11.84LKK500 pKa = 10.46NRR502 pKa = 11.84LKK504 pKa = 9.62TRR506 pKa = 11.84VRR508 pKa = 11.84TRR510 pKa = 11.84LKK512 pKa = 9.79TRR514 pKa = 11.84LKK516 pKa = 9.71TRR518 pKa = 11.84LKK520 pKa = 9.71TRR522 pKa = 11.84LKK524 pKa = 9.71TRR526 pKa = 11.84LKK528 pKa = 8.97TRR530 pKa = 11.84LRR532 pKa = 11.84TRR534 pKa = 11.84LKK536 pKa = 9.57TRR538 pKa = 11.84LKK540 pKa = 10.74NRR542 pKa = 11.84LKK544 pKa = 10.09TRR546 pKa = 11.84LKK548 pKa = 9.65TRR550 pKa = 11.84LKK552 pKa = 9.71TRR554 pKa = 11.84LKK556 pKa = 8.97TRR558 pKa = 11.84LRR560 pKa = 11.84TRR562 pKa = 11.84LKK564 pKa = 10.71NRR566 pKa = 11.84LKK568 pKa = 10.09TRR570 pKa = 11.84LKK572 pKa = 9.65TRR574 pKa = 11.84LKK576 pKa = 9.38TRR578 pKa = 11.84LKK580 pKa = 10.87ARR582 pKa = 11.84LKK584 pKa = 9.77VAFISPHH591 pKa = 5.36VEE593 pKa = 3.7EE594 pKa = 4.6TASSRR599 pKa = 11.84RR600 pKa = 11.84KK601 pKa = 10.39EE602 pKa = 3.69EE603 pKa = 4.01DD604 pKa = 2.77
Molecular weight: 75.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
33321467 |
17 |
7540 |
597.8 |
66.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.778 ± 0.009 | 2.313 ± 0.007 |
5.267 ± 0.008 | 6.722 ± 0.012 |
3.903 ± 0.007 | 6.412 ± 0.01 |
2.642 ± 0.005 | 4.418 ± 0.007 |
5.479 ± 0.01 | 9.784 ± 0.013 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.323 ± 0.004 | 3.738 ± 0.006 |
5.566 ± 0.012 | 4.58 ± 0.008 |
5.788 ± 0.008 | 8.345 ± 0.013 |
5.386 ± 0.009 | 6.524 ± 0.008 |
1.205 ± 0.003 | 2.826 ± 0.005 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
