
Kiritimatiella glycovorans
Taxonomy: cellular organisms; Bacteria; PVC group; Kiritimatiellaeota; Kiritimatiellae; Kiritimatiellales; Kiritimatiellaceae; Kiritimatiella
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
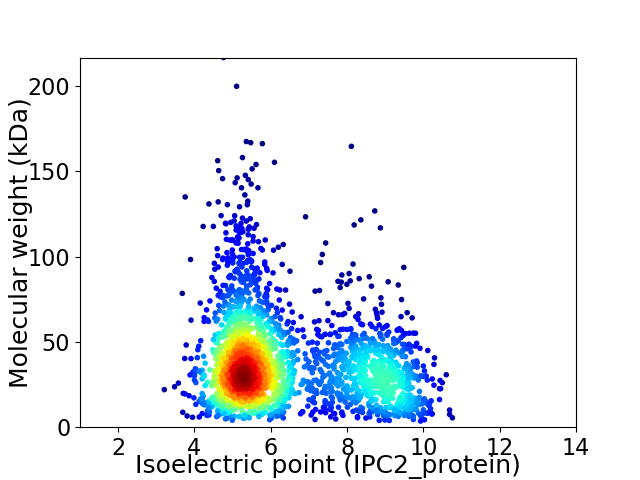
Virtual 2D-PAGE plot for 2373 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G3EGF0|A0A0G3EGF0_9BACT Putative fructose-bisphosphate aldolase OS=Kiritimatiella glycovorans OX=1307763 GN=fbaA PE=4 SV=1
MM1 pKa = 7.75RR2 pKa = 11.84AFFSAACFTCLAFGAVSAHH21 pKa = 5.52AVSVTTTSLPTGAVAEE37 pKa = 4.85AYY39 pKa = 10.45SGTLSASGGTTPYY52 pKa = 9.95RR53 pKa = 11.84WNVAQDD59 pKa = 3.59YY60 pKa = 11.03AEE62 pKa = 4.62SIEE65 pKa = 4.34TNAMQDD71 pKa = 3.3PSADD75 pKa = 3.62MDD77 pKa = 3.69WNADD81 pKa = 3.48SEE83 pKa = 4.63AWLLDD88 pKa = 3.56LPFTFPFYY96 pKa = 11.01GVGYY100 pKa = 5.11TQCWVAVDD108 pKa = 4.91GYY110 pKa = 10.13VHH112 pKa = 7.37FATNYY117 pKa = 10.27LDD119 pKa = 3.74STPTTNEE126 pKa = 3.56LAQQCMIAADD136 pKa = 3.93WKK138 pKa = 11.24DD139 pKa = 3.57LDD141 pKa = 3.93TSAGGDD147 pKa = 3.42LDD149 pKa = 3.9IYY151 pKa = 10.74IWTNDD156 pKa = 3.31SSCVIRR162 pKa = 11.84WYY164 pKa = 10.81GRR166 pKa = 11.84TPMWQYY172 pKa = 11.76VGIKK176 pKa = 10.5LEE178 pKa = 4.11MNEE181 pKa = 4.04NGLIRR186 pKa = 11.84VDD188 pKa = 3.53MSGDD192 pKa = 3.31EE193 pKa = 4.64GMMSPRR199 pKa = 11.84FCGLSAGNGVDD210 pKa = 3.8YY211 pKa = 11.13VVSTNNDD218 pKa = 2.47AWLDD222 pKa = 3.65PGDD225 pKa = 4.14IIRR228 pKa = 11.84FQPEE232 pKa = 3.63DD233 pKa = 3.76ALPTNIVMTSGGEE246 pKa = 4.06LYY248 pKa = 8.13GTPWNAGTNSLTFRR262 pKa = 11.84VTDD265 pKa = 3.53ATNGQAAAALDD276 pKa = 3.81LVVTEE281 pKa = 5.07GGNAKK286 pKa = 9.82PVVSGSTPAAGAYY299 pKa = 9.83PMDD302 pKa = 3.98EE303 pKa = 4.65AEE305 pKa = 4.39TNTFSVAASDD315 pKa = 4.11PDD317 pKa = 4.05GTPLSYY323 pKa = 11.09AWTLDD328 pKa = 3.22GTAVGTNSASYY339 pKa = 11.25DD340 pKa = 3.41FGTPWGGAGVYY351 pKa = 9.01TLLVEE356 pKa = 4.69ISDD359 pKa = 4.62DD360 pKa = 3.33FWTNGEE366 pKa = 4.36VSVEE370 pKa = 3.7WTVAVTNDD378 pKa = 3.13NDD380 pKa = 4.14GDD382 pKa = 3.88GMLNEE387 pKa = 4.21WEE389 pKa = 4.16RR390 pKa = 11.84TYY392 pKa = 11.72GLDD395 pKa = 4.52PEE397 pKa = 5.02VNDD400 pKa = 4.85SGGDD404 pKa = 3.51GDD406 pKa = 6.01GDD408 pKa = 3.58GLSNIEE414 pKa = 4.01EE415 pKa = 4.41HH416 pKa = 7.66DD417 pKa = 4.0AGTDD421 pKa = 3.47PGSEE425 pKa = 4.03DD426 pKa = 3.49TDD428 pKa = 3.61NDD430 pKa = 3.76TLYY433 pKa = 11.24DD434 pKa = 3.9DD435 pKa = 4.22WEE437 pKa = 4.63LEE439 pKa = 4.02YY440 pKa = 10.75GYY442 pKa = 10.9DD443 pKa = 3.44PTSPSEE449 pKa = 3.97ILPGYY454 pKa = 8.21TNYY457 pKa = 9.8TPSYY461 pKa = 9.53GYY463 pKa = 10.52NLSADD468 pKa = 3.61AEE470 pKa = 4.39EE471 pKa = 5.24VYY473 pKa = 9.42VTGTTAYY480 pKa = 9.43VACDD484 pKa = 3.28VGGLKK489 pKa = 10.68VIDD492 pKa = 4.98CSDD495 pKa = 3.54PDD497 pKa = 4.11APVLIDD503 pKa = 3.13MHH505 pKa = 6.81EE506 pKa = 4.04IPGRR510 pKa = 11.84AEE512 pKa = 4.31EE513 pKa = 4.18ICGLGPYY520 pKa = 10.08VFVGADD526 pKa = 3.31NYY528 pKa = 11.59GLTIFDD534 pKa = 4.59ASTPSNLAVLCTWTNAHH551 pKa = 7.02LFYY554 pKa = 11.22GPMAAVGDD562 pKa = 4.15RR563 pKa = 11.84LYY565 pKa = 11.06VAQNSDD571 pKa = 3.78LTVLSISDD579 pKa = 3.76PASPVLLGQLDD590 pKa = 3.96VSNTNHH596 pKa = 7.36PAPGSIRR603 pKa = 11.84DD604 pKa = 3.47VAVRR608 pKa = 11.84GDD610 pKa = 3.56YY611 pKa = 10.47AYY613 pKa = 10.72CANFTEE619 pKa = 4.61GLKK622 pKa = 9.34TVNISDD628 pKa = 4.5PEE630 pKa = 4.44TMVITATNEE639 pKa = 3.81YY640 pKa = 9.76EE641 pKa = 4.29GVGVCEE647 pKa = 3.94YY648 pKa = 10.79AARR651 pKa = 11.84GLDD654 pKa = 3.49IEE656 pKa = 5.05GDD658 pKa = 4.09TLFMCQDD665 pKa = 3.76GYY667 pKa = 12.0GLMAFDD673 pKa = 5.48LSDD676 pKa = 3.91PASPALIDD684 pKa = 4.21NFISSHH690 pKa = 6.15NPFPYY695 pKa = 10.53DD696 pKa = 3.24NLDD699 pKa = 3.81DD700 pKa = 4.28VDD702 pKa = 4.48VEE704 pKa = 4.38GTNAYY709 pKa = 10.25VIGGKK714 pKa = 9.62RR715 pKa = 11.84FWIYY719 pKa = 10.86GVSDD723 pKa = 4.13PSNMVEE729 pKa = 4.15LAHH732 pKa = 6.03WQNGSGGPTDD742 pKa = 4.98GIGIHH747 pKa = 7.09DD748 pKa = 4.22VDD750 pKa = 4.18AEE752 pKa = 4.38GDD754 pKa = 4.18FVHH757 pKa = 7.09LPTGWGTYY765 pKa = 7.36TNAYY769 pKa = 9.15GIQVDD774 pKa = 3.73RR775 pKa = 11.84GGLRR779 pKa = 11.84LMDD782 pKa = 4.09VTDD785 pKa = 3.85KK786 pKa = 11.13AAPEE790 pKa = 3.67PRR792 pKa = 11.84GRR794 pKa = 11.84FITSGQGWDD803 pKa = 3.46MEE805 pKa = 4.34ILNDD809 pKa = 3.37VAYY812 pKa = 10.1VANYY816 pKa = 9.25HH817 pKa = 6.02SGVHH821 pKa = 5.48IVNVTNWTRR830 pKa = 11.84LGRR833 pKa = 11.84YY834 pKa = 6.49DD835 pKa = 3.55TEE837 pKa = 5.15GIAMGIDD844 pKa = 3.45VTDD847 pKa = 4.02GGDD850 pKa = 3.38TLLVADD856 pKa = 5.06GEE858 pKa = 4.5NGLVVLDD865 pKa = 4.04ATDD868 pKa = 4.08PANMALQSHH877 pKa = 7.45LDD879 pKa = 3.55TDD881 pKa = 3.68DD882 pKa = 3.65ARR884 pKa = 11.84AVRR887 pKa = 11.84YY888 pKa = 9.83LNGRR892 pKa = 11.84IYY894 pKa = 10.8VADD897 pKa = 3.8YY898 pKa = 11.22TNGLLVATNLSVPTLQTNFFAGSPIQGLTTYY929 pKa = 10.44GSKK932 pKa = 10.61VYY934 pKa = 10.37AAAGGLGVYY943 pKa = 9.52SVYY946 pKa = 10.83CEE948 pKa = 3.82EE949 pKa = 4.59KK950 pKa = 10.46GSNEE954 pKa = 3.77TWHH957 pKa = 6.36TEE959 pKa = 3.65HH960 pKa = 6.76YY961 pKa = 10.7NYY963 pKa = 9.75TIDD966 pKa = 3.51MGKK969 pKa = 9.58SASAYY974 pKa = 9.81RR975 pKa = 11.84LEE977 pKa = 4.45AADD980 pKa = 3.42GRR982 pKa = 11.84LWVADD987 pKa = 3.57NLNGVVAIDD996 pKa = 3.59ISSAAMSYY1004 pKa = 10.62EE1005 pKa = 3.63GLYY1008 pKa = 10.24TNGVVQNVFIEE1019 pKa = 4.23DD1020 pKa = 3.61HH1021 pKa = 6.94APTGDD1026 pKa = 4.02YY1027 pKa = 10.42IWIGGGTNITSFRR1040 pKa = 11.84VGDD1043 pKa = 4.36PIDD1046 pKa = 3.76RR1047 pKa = 11.84RR1048 pKa = 11.84NTMSDD1053 pKa = 3.63EE1054 pKa = 4.3YY1055 pKa = 9.87WWQDD1059 pKa = 3.77PEE1061 pKa = 5.87IKK1063 pKa = 10.41DD1064 pKa = 3.28LWFAGTNGIFLGATQWKK1081 pKa = 9.1KK1082 pKa = 10.62YY1083 pKa = 10.03EE1084 pKa = 3.83YY1085 pKa = 10.78GLRR1088 pKa = 11.84SFNTTRR1094 pKa = 11.84VDD1096 pKa = 3.39ADD1098 pKa = 3.72EE1099 pKa = 6.63DD1100 pKa = 3.94GMADD1104 pKa = 3.58SDD1106 pKa = 3.84EE1107 pKa = 4.12MRR1109 pKa = 11.84WWGSTVPGRR1118 pKa = 11.84YY1119 pKa = 9.47DD1120 pKa = 3.31DD1121 pKa = 3.75TDD1123 pKa = 4.41GDD1125 pKa = 4.28GLSNWGEE1132 pKa = 4.38STTGTDD1138 pKa = 4.17PSLTDD1143 pKa = 3.18TDD1145 pKa = 4.01GDD1147 pKa = 4.25GVSDD1151 pKa = 3.81RR1152 pKa = 11.84DD1153 pKa = 3.62EE1154 pKa = 4.91YY1155 pKa = 11.09IAGTDD1160 pKa = 3.54PMSDD1164 pKa = 3.88GSCFSMDD1171 pKa = 3.83LDD1173 pKa = 4.22DD1174 pKa = 5.54PVYY1177 pKa = 10.78HH1178 pKa = 7.35LDD1180 pKa = 3.9LFDD1183 pKa = 4.32EE1184 pKa = 5.65FVLRR1188 pKa = 11.84WPSRR1192 pKa = 11.84AGRR1195 pKa = 11.84VYY1197 pKa = 11.15DD1198 pKa = 3.81LYY1200 pKa = 10.85RR1201 pKa = 11.84YY1202 pKa = 10.17SDD1204 pKa = 3.85LGDD1207 pKa = 3.59PSSRR1211 pKa = 11.84VQLLTNAPATPPMNIYY1227 pKa = 9.3TDD1229 pKa = 3.45TVFRR1233 pKa = 11.84IRR1235 pKa = 11.84SYY1237 pKa = 9.09MYY1239 pKa = 9.38RR1240 pKa = 11.84INVEE1244 pKa = 3.99RR1245 pKa = 11.84EE1246 pKa = 3.67
MM1 pKa = 7.75RR2 pKa = 11.84AFFSAACFTCLAFGAVSAHH21 pKa = 5.52AVSVTTTSLPTGAVAEE37 pKa = 4.85AYY39 pKa = 10.45SGTLSASGGTTPYY52 pKa = 9.95RR53 pKa = 11.84WNVAQDD59 pKa = 3.59YY60 pKa = 11.03AEE62 pKa = 4.62SIEE65 pKa = 4.34TNAMQDD71 pKa = 3.3PSADD75 pKa = 3.62MDD77 pKa = 3.69WNADD81 pKa = 3.48SEE83 pKa = 4.63AWLLDD88 pKa = 3.56LPFTFPFYY96 pKa = 11.01GVGYY100 pKa = 5.11TQCWVAVDD108 pKa = 4.91GYY110 pKa = 10.13VHH112 pKa = 7.37FATNYY117 pKa = 10.27LDD119 pKa = 3.74STPTTNEE126 pKa = 3.56LAQQCMIAADD136 pKa = 3.93WKK138 pKa = 11.24DD139 pKa = 3.57LDD141 pKa = 3.93TSAGGDD147 pKa = 3.42LDD149 pKa = 3.9IYY151 pKa = 10.74IWTNDD156 pKa = 3.31SSCVIRR162 pKa = 11.84WYY164 pKa = 10.81GRR166 pKa = 11.84TPMWQYY172 pKa = 11.76VGIKK176 pKa = 10.5LEE178 pKa = 4.11MNEE181 pKa = 4.04NGLIRR186 pKa = 11.84VDD188 pKa = 3.53MSGDD192 pKa = 3.31EE193 pKa = 4.64GMMSPRR199 pKa = 11.84FCGLSAGNGVDD210 pKa = 3.8YY211 pKa = 11.13VVSTNNDD218 pKa = 2.47AWLDD222 pKa = 3.65PGDD225 pKa = 4.14IIRR228 pKa = 11.84FQPEE232 pKa = 3.63DD233 pKa = 3.76ALPTNIVMTSGGEE246 pKa = 4.06LYY248 pKa = 8.13GTPWNAGTNSLTFRR262 pKa = 11.84VTDD265 pKa = 3.53ATNGQAAAALDD276 pKa = 3.81LVVTEE281 pKa = 5.07GGNAKK286 pKa = 9.82PVVSGSTPAAGAYY299 pKa = 9.83PMDD302 pKa = 3.98EE303 pKa = 4.65AEE305 pKa = 4.39TNTFSVAASDD315 pKa = 4.11PDD317 pKa = 4.05GTPLSYY323 pKa = 11.09AWTLDD328 pKa = 3.22GTAVGTNSASYY339 pKa = 11.25DD340 pKa = 3.41FGTPWGGAGVYY351 pKa = 9.01TLLVEE356 pKa = 4.69ISDD359 pKa = 4.62DD360 pKa = 3.33FWTNGEE366 pKa = 4.36VSVEE370 pKa = 3.7WTVAVTNDD378 pKa = 3.13NDD380 pKa = 4.14GDD382 pKa = 3.88GMLNEE387 pKa = 4.21WEE389 pKa = 4.16RR390 pKa = 11.84TYY392 pKa = 11.72GLDD395 pKa = 4.52PEE397 pKa = 5.02VNDD400 pKa = 4.85SGGDD404 pKa = 3.51GDD406 pKa = 6.01GDD408 pKa = 3.58GLSNIEE414 pKa = 4.01EE415 pKa = 4.41HH416 pKa = 7.66DD417 pKa = 4.0AGTDD421 pKa = 3.47PGSEE425 pKa = 4.03DD426 pKa = 3.49TDD428 pKa = 3.61NDD430 pKa = 3.76TLYY433 pKa = 11.24DD434 pKa = 3.9DD435 pKa = 4.22WEE437 pKa = 4.63LEE439 pKa = 4.02YY440 pKa = 10.75GYY442 pKa = 10.9DD443 pKa = 3.44PTSPSEE449 pKa = 3.97ILPGYY454 pKa = 8.21TNYY457 pKa = 9.8TPSYY461 pKa = 9.53GYY463 pKa = 10.52NLSADD468 pKa = 3.61AEE470 pKa = 4.39EE471 pKa = 5.24VYY473 pKa = 9.42VTGTTAYY480 pKa = 9.43VACDD484 pKa = 3.28VGGLKK489 pKa = 10.68VIDD492 pKa = 4.98CSDD495 pKa = 3.54PDD497 pKa = 4.11APVLIDD503 pKa = 3.13MHH505 pKa = 6.81EE506 pKa = 4.04IPGRR510 pKa = 11.84AEE512 pKa = 4.31EE513 pKa = 4.18ICGLGPYY520 pKa = 10.08VFVGADD526 pKa = 3.31NYY528 pKa = 11.59GLTIFDD534 pKa = 4.59ASTPSNLAVLCTWTNAHH551 pKa = 7.02LFYY554 pKa = 11.22GPMAAVGDD562 pKa = 4.15RR563 pKa = 11.84LYY565 pKa = 11.06VAQNSDD571 pKa = 3.78LTVLSISDD579 pKa = 3.76PASPVLLGQLDD590 pKa = 3.96VSNTNHH596 pKa = 7.36PAPGSIRR603 pKa = 11.84DD604 pKa = 3.47VAVRR608 pKa = 11.84GDD610 pKa = 3.56YY611 pKa = 10.47AYY613 pKa = 10.72CANFTEE619 pKa = 4.61GLKK622 pKa = 9.34TVNISDD628 pKa = 4.5PEE630 pKa = 4.44TMVITATNEE639 pKa = 3.81YY640 pKa = 9.76EE641 pKa = 4.29GVGVCEE647 pKa = 3.94YY648 pKa = 10.79AARR651 pKa = 11.84GLDD654 pKa = 3.49IEE656 pKa = 5.05GDD658 pKa = 4.09TLFMCQDD665 pKa = 3.76GYY667 pKa = 12.0GLMAFDD673 pKa = 5.48LSDD676 pKa = 3.91PASPALIDD684 pKa = 4.21NFISSHH690 pKa = 6.15NPFPYY695 pKa = 10.53DD696 pKa = 3.24NLDD699 pKa = 3.81DD700 pKa = 4.28VDD702 pKa = 4.48VEE704 pKa = 4.38GTNAYY709 pKa = 10.25VIGGKK714 pKa = 9.62RR715 pKa = 11.84FWIYY719 pKa = 10.86GVSDD723 pKa = 4.13PSNMVEE729 pKa = 4.15LAHH732 pKa = 6.03WQNGSGGPTDD742 pKa = 4.98GIGIHH747 pKa = 7.09DD748 pKa = 4.22VDD750 pKa = 4.18AEE752 pKa = 4.38GDD754 pKa = 4.18FVHH757 pKa = 7.09LPTGWGTYY765 pKa = 7.36TNAYY769 pKa = 9.15GIQVDD774 pKa = 3.73RR775 pKa = 11.84GGLRR779 pKa = 11.84LMDD782 pKa = 4.09VTDD785 pKa = 3.85KK786 pKa = 11.13AAPEE790 pKa = 3.67PRR792 pKa = 11.84GRR794 pKa = 11.84FITSGQGWDD803 pKa = 3.46MEE805 pKa = 4.34ILNDD809 pKa = 3.37VAYY812 pKa = 10.1VANYY816 pKa = 9.25HH817 pKa = 6.02SGVHH821 pKa = 5.48IVNVTNWTRR830 pKa = 11.84LGRR833 pKa = 11.84YY834 pKa = 6.49DD835 pKa = 3.55TEE837 pKa = 5.15GIAMGIDD844 pKa = 3.45VTDD847 pKa = 4.02GGDD850 pKa = 3.38TLLVADD856 pKa = 5.06GEE858 pKa = 4.5NGLVVLDD865 pKa = 4.04ATDD868 pKa = 4.08PANMALQSHH877 pKa = 7.45LDD879 pKa = 3.55TDD881 pKa = 3.68DD882 pKa = 3.65ARR884 pKa = 11.84AVRR887 pKa = 11.84YY888 pKa = 9.83LNGRR892 pKa = 11.84IYY894 pKa = 10.8VADD897 pKa = 3.8YY898 pKa = 11.22TNGLLVATNLSVPTLQTNFFAGSPIQGLTTYY929 pKa = 10.44GSKK932 pKa = 10.61VYY934 pKa = 10.37AAAGGLGVYY943 pKa = 9.52SVYY946 pKa = 10.83CEE948 pKa = 3.82EE949 pKa = 4.59KK950 pKa = 10.46GSNEE954 pKa = 3.77TWHH957 pKa = 6.36TEE959 pKa = 3.65HH960 pKa = 6.76YY961 pKa = 10.7NYY963 pKa = 9.75TIDD966 pKa = 3.51MGKK969 pKa = 9.58SASAYY974 pKa = 9.81RR975 pKa = 11.84LEE977 pKa = 4.45AADD980 pKa = 3.42GRR982 pKa = 11.84LWVADD987 pKa = 3.57NLNGVVAIDD996 pKa = 3.59ISSAAMSYY1004 pKa = 10.62EE1005 pKa = 3.63GLYY1008 pKa = 10.24TNGVVQNVFIEE1019 pKa = 4.23DD1020 pKa = 3.61HH1021 pKa = 6.94APTGDD1026 pKa = 4.02YY1027 pKa = 10.42IWIGGGTNITSFRR1040 pKa = 11.84VGDD1043 pKa = 4.36PIDD1046 pKa = 3.76RR1047 pKa = 11.84RR1048 pKa = 11.84NTMSDD1053 pKa = 3.63EE1054 pKa = 4.3YY1055 pKa = 9.87WWQDD1059 pKa = 3.77PEE1061 pKa = 5.87IKK1063 pKa = 10.41DD1064 pKa = 3.28LWFAGTNGIFLGATQWKK1081 pKa = 9.1KK1082 pKa = 10.62YY1083 pKa = 10.03EE1084 pKa = 3.83YY1085 pKa = 10.78GLRR1088 pKa = 11.84SFNTTRR1094 pKa = 11.84VDD1096 pKa = 3.39ADD1098 pKa = 3.72EE1099 pKa = 6.63DD1100 pKa = 3.94GMADD1104 pKa = 3.58SDD1106 pKa = 3.84EE1107 pKa = 4.12MRR1109 pKa = 11.84WWGSTVPGRR1118 pKa = 11.84YY1119 pKa = 9.47DD1120 pKa = 3.31DD1121 pKa = 3.75TDD1123 pKa = 4.41GDD1125 pKa = 4.28GLSNWGEE1132 pKa = 4.38STTGTDD1138 pKa = 4.17PSLTDD1143 pKa = 3.18TDD1145 pKa = 4.01GDD1147 pKa = 4.25GVSDD1151 pKa = 3.81RR1152 pKa = 11.84DD1153 pKa = 3.62EE1154 pKa = 4.91YY1155 pKa = 11.09IAGTDD1160 pKa = 3.54PMSDD1164 pKa = 3.88GSCFSMDD1171 pKa = 3.83LDD1173 pKa = 4.22DD1174 pKa = 5.54PVYY1177 pKa = 10.78HH1178 pKa = 7.35LDD1180 pKa = 3.9LFDD1183 pKa = 4.32EE1184 pKa = 5.65FVLRR1188 pKa = 11.84WPSRR1192 pKa = 11.84AGRR1195 pKa = 11.84VYY1197 pKa = 11.15DD1198 pKa = 3.81LYY1200 pKa = 10.85RR1201 pKa = 11.84YY1202 pKa = 10.17SDD1204 pKa = 3.85LGDD1207 pKa = 3.59PSSRR1211 pKa = 11.84VQLLTNAPATPPMNIYY1227 pKa = 9.3TDD1229 pKa = 3.45TVFRR1233 pKa = 11.84IRR1235 pKa = 11.84SYY1237 pKa = 9.09MYY1239 pKa = 9.38RR1240 pKa = 11.84INVEE1244 pKa = 3.99RR1245 pKa = 11.84EE1246 pKa = 3.67
Molecular weight: 134.99 kDa
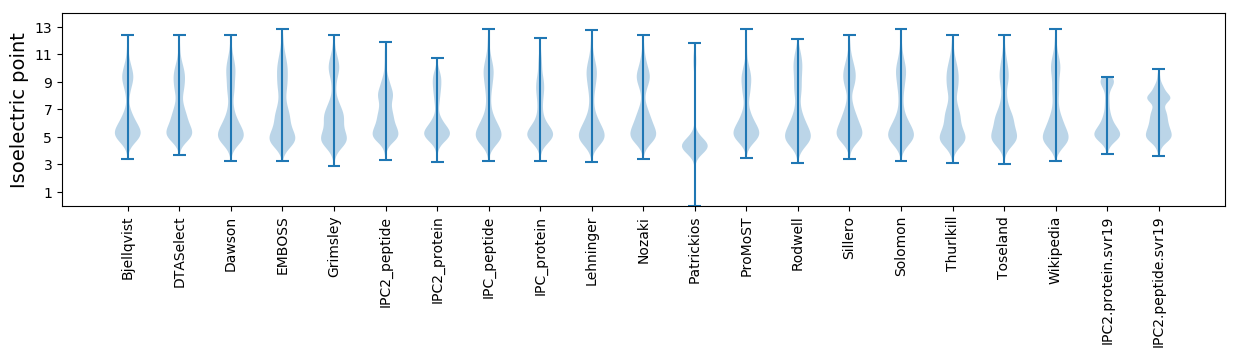
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G3ED69|A0A0G3ED69_9BACT Twitching mobility protein OS=Kiritimatiella glycovorans OX=1307763 GN=pilT_2 PE=3 SV=1
MM1 pKa = 7.4IAGGRR6 pKa = 11.84IQVNGEE12 pKa = 4.32TVCEE16 pKa = 3.83QGIKK20 pKa = 9.97VDD22 pKa = 4.11PVQDD26 pKa = 4.67DD27 pKa = 2.77IRR29 pKa = 11.84MDD31 pKa = 3.17GRR33 pKa = 11.84VVRR36 pKa = 11.84RR37 pKa = 11.84EE38 pKa = 3.99SKK40 pKa = 10.32VVYY43 pKa = 10.06ILNKK47 pKa = 9.56PPGVMCTNDD56 pKa = 3.92DD57 pKa = 3.75PQSRR61 pKa = 11.84PLYY64 pKa = 9.97RR65 pKa = 11.84DD66 pKa = 4.28LFPADD71 pKa = 3.63AGRR74 pKa = 11.84LFPVGRR80 pKa = 11.84LDD82 pKa = 4.48CMSEE86 pKa = 3.87GLLLVTNDD94 pKa = 2.63GDD96 pKa = 3.87LAHH99 pKa = 7.42RR100 pKa = 11.84LMHH103 pKa = 6.29PRR105 pKa = 11.84YY106 pKa = 9.09GFHH109 pKa = 6.19KK110 pKa = 8.95TYY112 pKa = 10.6LARR115 pKa = 11.84TPDD118 pKa = 3.43RR119 pKa = 11.84VTAEE123 pKa = 3.47QMKK126 pKa = 10.42RR127 pKa = 11.84MEE129 pKa = 4.19QGVEE133 pKa = 3.8EE134 pKa = 4.53GGEE137 pKa = 3.88RR138 pKa = 11.84LRR140 pKa = 11.84ALRR143 pKa = 11.84VRR145 pKa = 11.84RR146 pKa = 11.84EE147 pKa = 3.76RR148 pKa = 11.84GAGPGTRR155 pKa = 11.84LRR157 pKa = 11.84IVLGEE162 pKa = 3.61GRR164 pKa = 11.84NRR166 pKa = 11.84HH167 pKa = 4.64IRR169 pKa = 11.84RR170 pKa = 11.84MFKK173 pKa = 10.71ALGLRR178 pKa = 11.84IDD180 pKa = 3.35RR181 pKa = 11.84LKK183 pKa = 10.63RR184 pKa = 11.84VKK186 pKa = 10.51LGPLVLGGLPQGSWRR201 pKa = 11.84EE202 pKa = 3.86LRR204 pKa = 11.84PQEE207 pKa = 4.28LRR209 pKa = 11.84QLQVSSHH216 pKa = 5.17QRR218 pKa = 11.84DD219 pKa = 3.27AA220 pKa = 4.25
MM1 pKa = 7.4IAGGRR6 pKa = 11.84IQVNGEE12 pKa = 4.32TVCEE16 pKa = 3.83QGIKK20 pKa = 9.97VDD22 pKa = 4.11PVQDD26 pKa = 4.67DD27 pKa = 2.77IRR29 pKa = 11.84MDD31 pKa = 3.17GRR33 pKa = 11.84VVRR36 pKa = 11.84RR37 pKa = 11.84EE38 pKa = 3.99SKK40 pKa = 10.32VVYY43 pKa = 10.06ILNKK47 pKa = 9.56PPGVMCTNDD56 pKa = 3.92DD57 pKa = 3.75PQSRR61 pKa = 11.84PLYY64 pKa = 9.97RR65 pKa = 11.84DD66 pKa = 4.28LFPADD71 pKa = 3.63AGRR74 pKa = 11.84LFPVGRR80 pKa = 11.84LDD82 pKa = 4.48CMSEE86 pKa = 3.87GLLLVTNDD94 pKa = 2.63GDD96 pKa = 3.87LAHH99 pKa = 7.42RR100 pKa = 11.84LMHH103 pKa = 6.29PRR105 pKa = 11.84YY106 pKa = 9.09GFHH109 pKa = 6.19KK110 pKa = 8.95TYY112 pKa = 10.6LARR115 pKa = 11.84TPDD118 pKa = 3.43RR119 pKa = 11.84VTAEE123 pKa = 3.47QMKK126 pKa = 10.42RR127 pKa = 11.84MEE129 pKa = 4.19QGVEE133 pKa = 3.8EE134 pKa = 4.53GGEE137 pKa = 3.88RR138 pKa = 11.84LRR140 pKa = 11.84ALRR143 pKa = 11.84VRR145 pKa = 11.84RR146 pKa = 11.84EE147 pKa = 3.76RR148 pKa = 11.84GAGPGTRR155 pKa = 11.84LRR157 pKa = 11.84IVLGEE162 pKa = 3.61GRR164 pKa = 11.84NRR166 pKa = 11.84HH167 pKa = 4.64IRR169 pKa = 11.84RR170 pKa = 11.84MFKK173 pKa = 10.71ALGLRR178 pKa = 11.84IDD180 pKa = 3.35RR181 pKa = 11.84LKK183 pKa = 10.63RR184 pKa = 11.84VKK186 pKa = 10.51LGPLVLGGLPQGSWRR201 pKa = 11.84EE202 pKa = 3.86LRR204 pKa = 11.84PQEE207 pKa = 4.28LRR209 pKa = 11.84QLQVSSHH216 pKa = 5.17QRR218 pKa = 11.84DD219 pKa = 3.27AA220 pKa = 4.25
Molecular weight: 25.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
860530 |
29 |
1991 |
362.6 |
40.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.122 ± 0.054 | 1.255 ± 0.018 |
6.006 ± 0.045 | 7.464 ± 0.048 |
3.814 ± 0.027 | 8.514 ± 0.045 |
2.348 ± 0.024 | 4.606 ± 0.033 |
3.111 ± 0.042 | 9.621 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.411 ± 0.022 | 2.636 ± 0.033 |
5.265 ± 0.036 | 2.697 ± 0.027 |
8.854 ± 0.055 | 5.058 ± 0.034 |
4.965 ± 0.037 | 6.95 ± 0.039 |
1.496 ± 0.024 | 2.807 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
