
Escherichia virus Qbeta (Bacteriophage Q-beta)
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Allassoviricetes; Levivirales; Leviviridae; Allolevivirus
Average proteome isoelectric point is 7.61
Get precalculated fractions of proteins
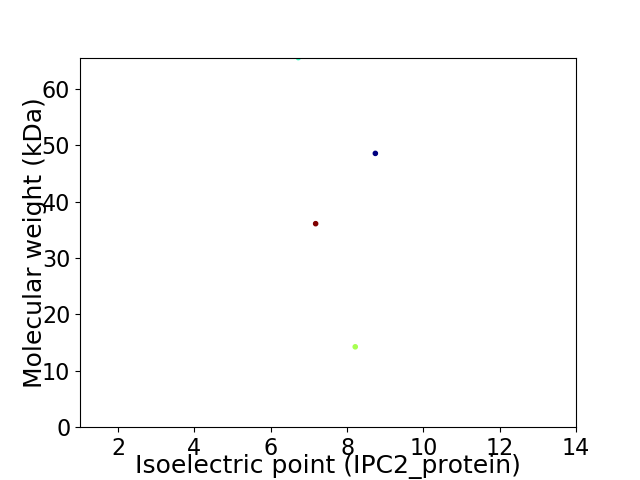
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
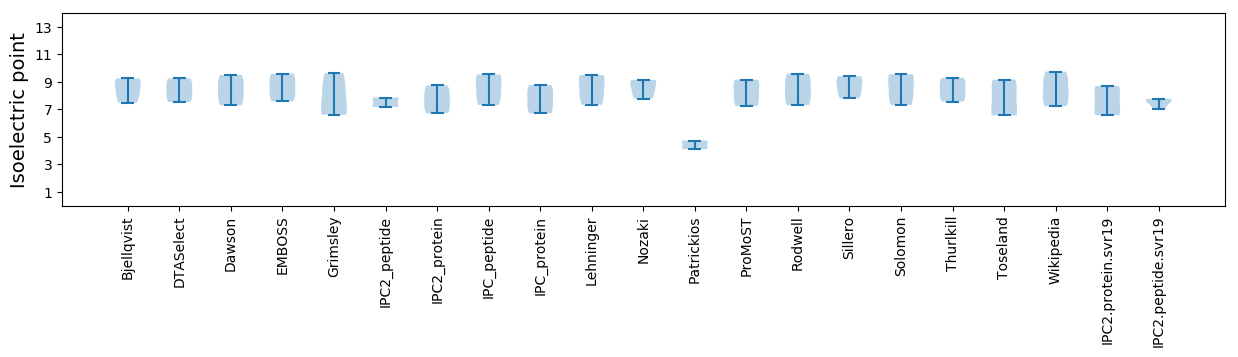
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q8LTE1|A1_BPQBE Minor capsid protein A1 OS=Escherichia virus Qbeta OX=39803 PE=1 SV=2
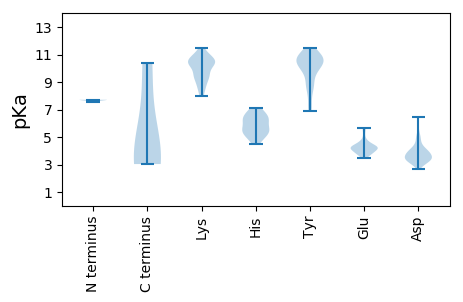
MM1 pKa = 7.56SKK3 pKa = 8.7TASSRR8 pKa = 11.84NSLSAQLRR16 pKa = 11.84RR17 pKa = 11.84AANTRR22 pKa = 11.84IEE24 pKa = 4.23VEE26 pKa = 4.16GNLALSIANDD36 pKa = 3.57LLLAYY41 pKa = 8.92GQSPFNSEE49 pKa = 4.07AEE51 pKa = 4.56CISFSPRR58 pKa = 11.84FDD60 pKa = 3.21GTPDD64 pKa = 3.48DD65 pKa = 5.01FRR67 pKa = 11.84INYY70 pKa = 8.89LKK72 pKa = 10.96AEE74 pKa = 4.19IMSKK78 pKa = 10.94YY79 pKa = 10.64DD80 pKa = 3.86DD81 pKa = 4.1FSLGIDD87 pKa = 3.6TEE89 pKa = 4.29AVAWEE94 pKa = 4.23KK95 pKa = 10.73FLAAEE100 pKa = 4.51AEE102 pKa = 4.45CALTNARR109 pKa = 11.84LYY111 pKa = 10.84RR112 pKa = 11.84PDD114 pKa = 3.36YY115 pKa = 11.43SEE117 pKa = 5.66DD118 pKa = 3.67FNFSLGEE125 pKa = 4.13SCIHH129 pKa = 5.12MARR132 pKa = 11.84RR133 pKa = 11.84KK134 pKa = 8.54IAKK137 pKa = 9.89LIGDD141 pKa = 3.94VPSVEE146 pKa = 3.85GMLRR150 pKa = 11.84HH151 pKa = 6.06CRR153 pKa = 11.84FSGGATTTNNRR164 pKa = 11.84SYY166 pKa = 11.02GHH168 pKa = 7.13PSFKK172 pKa = 10.34FALPQACTPRR182 pKa = 11.84ALKK185 pKa = 10.57YY186 pKa = 10.64VLALRR191 pKa = 11.84ASTHH195 pKa = 4.78FDD197 pKa = 2.69IRR199 pKa = 11.84ISDD202 pKa = 3.66ISPFNKK208 pKa = 10.01AVTVPKK214 pKa = 10.41NSKK217 pKa = 7.99TDD219 pKa = 3.07RR220 pKa = 11.84CIAIEE225 pKa = 4.3PGWNMFFQLGIGGILRR241 pKa = 11.84DD242 pKa = 4.47RR243 pKa = 11.84LRR245 pKa = 11.84CWGIDD250 pKa = 4.63LNDD253 pKa = 3.14QTINQRR259 pKa = 11.84RR260 pKa = 11.84AHH262 pKa = 6.4EE263 pKa = 4.41GSVTNNLATVDD274 pKa = 4.27LSAASDD280 pKa = 4.1SISLALCEE288 pKa = 4.79LLLPPGWFEE297 pKa = 4.03VLMDD301 pKa = 4.06LRR303 pKa = 11.84SPKK306 pKa = 10.13GRR308 pKa = 11.84LPDD311 pKa = 3.84GSVVTYY317 pKa = 9.8EE318 pKa = 5.13KK319 pKa = 10.24ISSMGNGYY327 pKa = 7.26TFEE330 pKa = 4.9LEE332 pKa = 4.21SLIFASLARR341 pKa = 11.84SVCEE345 pKa = 4.6ILDD348 pKa = 4.11LDD350 pKa = 3.97SSEE353 pKa = 4.05VTVYY357 pKa = 11.02GDD359 pKa = 5.34DD360 pKa = 4.8IILPSCAVPALRR372 pKa = 11.84EE373 pKa = 3.85VFKK376 pKa = 11.39YY377 pKa = 10.92VGFTTNTKK385 pKa = 8.63KK386 pKa = 9.18TFSEE390 pKa = 5.37GPFRR394 pKa = 11.84EE395 pKa = 4.49SCGKK399 pKa = 9.83HH400 pKa = 5.46YY401 pKa = 10.94YY402 pKa = 10.3SGVDD406 pKa = 3.12VTPFYY411 pKa = 10.39IRR413 pKa = 11.84HH414 pKa = 6.47RR415 pKa = 11.84IVSPADD421 pKa = 4.01LILVLNNLYY430 pKa = 10.17RR431 pKa = 11.84WATIDD436 pKa = 4.29GVWDD440 pKa = 3.98PRR442 pKa = 11.84AHH444 pKa = 6.3SVYY447 pKa = 10.64LKK449 pKa = 10.35YY450 pKa = 10.51RR451 pKa = 11.84KK452 pKa = 9.32LLPKK456 pKa = 9.88QLQRR460 pKa = 11.84NTIPDD465 pKa = 4.08GYY467 pKa = 11.09GDD469 pKa = 3.92GALVGSVLINPFAKK483 pKa = 9.42NRR485 pKa = 11.84GWIRR489 pKa = 11.84YY490 pKa = 7.93VPVITDD496 pKa = 3.28HH497 pKa = 5.71TRR499 pKa = 11.84DD500 pKa = 3.36RR501 pKa = 11.84EE502 pKa = 4.05RR503 pKa = 11.84AEE505 pKa = 4.27LGSYY509 pKa = 10.56LYY511 pKa = 10.85DD512 pKa = 3.66LFSRR516 pKa = 11.84CLSEE520 pKa = 5.64SNDD523 pKa = 3.45GLPLRR528 pKa = 11.84GPSGCDD534 pKa = 3.0SADD537 pKa = 3.31LFAIDD542 pKa = 5.07QLICRR547 pKa = 11.84SNPTKK552 pKa = 10.4ISRR555 pKa = 11.84STGKK559 pKa = 10.29FDD561 pKa = 3.18IQYY564 pKa = 8.74IACSSRR570 pKa = 11.84VLAPYY575 pKa = 10.57GVFQGTKK582 pKa = 9.16VASLHH587 pKa = 5.47EE588 pKa = 4.26AA589 pKa = 3.36
MM1 pKa = 7.56SKK3 pKa = 8.7TASSRR8 pKa = 11.84NSLSAQLRR16 pKa = 11.84RR17 pKa = 11.84AANTRR22 pKa = 11.84IEE24 pKa = 4.23VEE26 pKa = 4.16GNLALSIANDD36 pKa = 3.57LLLAYY41 pKa = 8.92GQSPFNSEE49 pKa = 4.07AEE51 pKa = 4.56CISFSPRR58 pKa = 11.84FDD60 pKa = 3.21GTPDD64 pKa = 3.48DD65 pKa = 5.01FRR67 pKa = 11.84INYY70 pKa = 8.89LKK72 pKa = 10.96AEE74 pKa = 4.19IMSKK78 pKa = 10.94YY79 pKa = 10.64DD80 pKa = 3.86DD81 pKa = 4.1FSLGIDD87 pKa = 3.6TEE89 pKa = 4.29AVAWEE94 pKa = 4.23KK95 pKa = 10.73FLAAEE100 pKa = 4.51AEE102 pKa = 4.45CALTNARR109 pKa = 11.84LYY111 pKa = 10.84RR112 pKa = 11.84PDD114 pKa = 3.36YY115 pKa = 11.43SEE117 pKa = 5.66DD118 pKa = 3.67FNFSLGEE125 pKa = 4.13SCIHH129 pKa = 5.12MARR132 pKa = 11.84RR133 pKa = 11.84KK134 pKa = 8.54IAKK137 pKa = 9.89LIGDD141 pKa = 3.94VPSVEE146 pKa = 3.85GMLRR150 pKa = 11.84HH151 pKa = 6.06CRR153 pKa = 11.84FSGGATTTNNRR164 pKa = 11.84SYY166 pKa = 11.02GHH168 pKa = 7.13PSFKK172 pKa = 10.34FALPQACTPRR182 pKa = 11.84ALKK185 pKa = 10.57YY186 pKa = 10.64VLALRR191 pKa = 11.84ASTHH195 pKa = 4.78FDD197 pKa = 2.69IRR199 pKa = 11.84ISDD202 pKa = 3.66ISPFNKK208 pKa = 10.01AVTVPKK214 pKa = 10.41NSKK217 pKa = 7.99TDD219 pKa = 3.07RR220 pKa = 11.84CIAIEE225 pKa = 4.3PGWNMFFQLGIGGILRR241 pKa = 11.84DD242 pKa = 4.47RR243 pKa = 11.84LRR245 pKa = 11.84CWGIDD250 pKa = 4.63LNDD253 pKa = 3.14QTINQRR259 pKa = 11.84RR260 pKa = 11.84AHH262 pKa = 6.4EE263 pKa = 4.41GSVTNNLATVDD274 pKa = 4.27LSAASDD280 pKa = 4.1SISLALCEE288 pKa = 4.79LLLPPGWFEE297 pKa = 4.03VLMDD301 pKa = 4.06LRR303 pKa = 11.84SPKK306 pKa = 10.13GRR308 pKa = 11.84LPDD311 pKa = 3.84GSVVTYY317 pKa = 9.8EE318 pKa = 5.13KK319 pKa = 10.24ISSMGNGYY327 pKa = 7.26TFEE330 pKa = 4.9LEE332 pKa = 4.21SLIFASLARR341 pKa = 11.84SVCEE345 pKa = 4.6ILDD348 pKa = 4.11LDD350 pKa = 3.97SSEE353 pKa = 4.05VTVYY357 pKa = 11.02GDD359 pKa = 5.34DD360 pKa = 4.8IILPSCAVPALRR372 pKa = 11.84EE373 pKa = 3.85VFKK376 pKa = 11.39YY377 pKa = 10.92VGFTTNTKK385 pKa = 8.63KK386 pKa = 9.18TFSEE390 pKa = 5.37GPFRR394 pKa = 11.84EE395 pKa = 4.49SCGKK399 pKa = 9.83HH400 pKa = 5.46YY401 pKa = 10.94YY402 pKa = 10.3SGVDD406 pKa = 3.12VTPFYY411 pKa = 10.39IRR413 pKa = 11.84HH414 pKa = 6.47RR415 pKa = 11.84IVSPADD421 pKa = 4.01LILVLNNLYY430 pKa = 10.17RR431 pKa = 11.84WATIDD436 pKa = 4.29GVWDD440 pKa = 3.98PRR442 pKa = 11.84AHH444 pKa = 6.3SVYY447 pKa = 10.64LKK449 pKa = 10.35YY450 pKa = 10.51RR451 pKa = 11.84KK452 pKa = 9.32LLPKK456 pKa = 9.88QLQRR460 pKa = 11.84NTIPDD465 pKa = 4.08GYY467 pKa = 11.09GDD469 pKa = 3.92GALVGSVLINPFAKK483 pKa = 9.42NRR485 pKa = 11.84GWIRR489 pKa = 11.84YY490 pKa = 7.93VPVITDD496 pKa = 3.28HH497 pKa = 5.71TRR499 pKa = 11.84DD500 pKa = 3.36RR501 pKa = 11.84EE502 pKa = 4.05RR503 pKa = 11.84AEE505 pKa = 4.27LGSYY509 pKa = 10.56LYY511 pKa = 10.85DD512 pKa = 3.66LFSRR516 pKa = 11.84CLSEE520 pKa = 5.64SNDD523 pKa = 3.45GLPLRR528 pKa = 11.84GPSGCDD534 pKa = 3.0SADD537 pKa = 3.31LFAIDD542 pKa = 5.07QLICRR547 pKa = 11.84SNPTKK552 pKa = 10.4ISRR555 pKa = 11.84STGKK559 pKa = 10.29FDD561 pKa = 3.18IQYY564 pKa = 8.74IACSSRR570 pKa = 11.84VLAPYY575 pKa = 10.57GVFQGTKK582 pKa = 9.16VASLHH587 pKa = 5.47EE588 pKa = 4.26AA589 pKa = 3.36
Molecular weight: 65.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q8LTE2|MATA2_BPQBE Maturation protein A2 OS=Escherichia virus Qbeta OX=39803 PE=1 SV=1
MM1 pKa = 7.71PKK3 pKa = 10.23LPRR6 pKa = 11.84GLRR9 pKa = 11.84FGADD13 pKa = 3.24NEE15 pKa = 4.22ILNDD19 pKa = 4.03FQEE22 pKa = 4.27LWFPDD27 pKa = 3.71LFIEE31 pKa = 5.64SSDD34 pKa = 3.31THH36 pKa = 5.46PWYY39 pKa = 9.22TLKK42 pKa = 10.9GRR44 pKa = 11.84VLNAHH49 pKa = 6.9LDD51 pKa = 3.64DD52 pKa = 4.99RR53 pKa = 11.84LPNVGGRR60 pKa = 11.84QVRR63 pKa = 11.84RR64 pKa = 11.84TPHH67 pKa = 6.39RR68 pKa = 11.84VTVPIASSGLRR79 pKa = 11.84PVTTVQYY86 pKa = 11.01DD87 pKa = 3.63PAALSFLLNARR98 pKa = 11.84VDD100 pKa = 3.28WDD102 pKa = 3.98FGNGDD107 pKa = 3.75SANLVINDD115 pKa = 3.19FLFRR119 pKa = 11.84TFAPKK124 pKa = 10.55EE125 pKa = 3.84FDD127 pKa = 3.51FSNSLVPRR135 pKa = 11.84YY136 pKa = 7.01TQAFSAFNAKK146 pKa = 9.28YY147 pKa = 8.72GTMIGEE153 pKa = 4.21GLEE156 pKa = 4.28TIKK159 pKa = 11.09YY160 pKa = 10.14LGLLLRR166 pKa = 11.84RR167 pKa = 11.84LRR169 pKa = 11.84EE170 pKa = 3.85GYY172 pKa = 9.97RR173 pKa = 11.84AVKK176 pKa = 10.45RR177 pKa = 11.84GDD179 pKa = 3.12LRR181 pKa = 11.84ALRR184 pKa = 11.84RR185 pKa = 11.84VIQSYY190 pKa = 11.34HH191 pKa = 5.03NGKK194 pKa = 8.62WKK196 pKa = 9.67PATAGNLWLEE206 pKa = 4.05FRR208 pKa = 11.84YY209 pKa = 10.88GLMPLFYY216 pKa = 10.44DD217 pKa = 3.88IRR219 pKa = 11.84DD220 pKa = 3.92VMLDD224 pKa = 3.02WQNRR228 pKa = 11.84HH229 pKa = 6.37DD230 pKa = 4.62KK231 pKa = 10.31IQRR234 pKa = 11.84LLRR237 pKa = 11.84FSVGHH242 pKa = 6.04GEE244 pKa = 4.23DD245 pKa = 3.78YY246 pKa = 11.19VVEE249 pKa = 4.32FDD251 pKa = 3.66NLYY254 pKa = 10.04PAVAYY259 pKa = 8.42FKK261 pKa = 11.07LKK263 pKa = 11.18GEE265 pKa = 3.9ITLEE269 pKa = 3.51RR270 pKa = 11.84RR271 pKa = 11.84HH272 pKa = 5.59RR273 pKa = 11.84HH274 pKa = 5.18GISYY278 pKa = 10.79ANRR281 pKa = 11.84EE282 pKa = 4.21GYY284 pKa = 10.74AVFDD288 pKa = 4.06NGSLRR293 pKa = 11.84PVSDD297 pKa = 3.0WKK299 pKa = 10.98EE300 pKa = 3.52LATAFINPHH309 pKa = 4.5EE310 pKa = 4.7VAWEE314 pKa = 3.73LTPYY318 pKa = 10.72SFVVDD323 pKa = 3.51WFLNVGDD330 pKa = 4.94ILAQQGQLYY339 pKa = 10.14HH340 pKa = 6.89NIDD343 pKa = 3.27IVDD346 pKa = 3.7GFDD349 pKa = 3.84RR350 pKa = 11.84RR351 pKa = 11.84DD352 pKa = 3.05IRR354 pKa = 11.84LKK356 pKa = 10.49SFTIKK361 pKa = 10.67GEE363 pKa = 4.24RR364 pKa = 11.84NGRR367 pKa = 11.84PVNVSASLSAVDD379 pKa = 4.34LFYY382 pKa = 11.51SRR384 pKa = 11.84LHH386 pKa = 5.8TSNLPFATLDD396 pKa = 3.44LDD398 pKa = 3.97TTFSSFKK405 pKa = 10.41HH406 pKa = 5.3VLDD409 pKa = 4.96SIFLLTQRR417 pKa = 11.84VKK419 pKa = 10.83RR420 pKa = 3.95
MM1 pKa = 7.71PKK3 pKa = 10.23LPRR6 pKa = 11.84GLRR9 pKa = 11.84FGADD13 pKa = 3.24NEE15 pKa = 4.22ILNDD19 pKa = 4.03FQEE22 pKa = 4.27LWFPDD27 pKa = 3.71LFIEE31 pKa = 5.64SSDD34 pKa = 3.31THH36 pKa = 5.46PWYY39 pKa = 9.22TLKK42 pKa = 10.9GRR44 pKa = 11.84VLNAHH49 pKa = 6.9LDD51 pKa = 3.64DD52 pKa = 4.99RR53 pKa = 11.84LPNVGGRR60 pKa = 11.84QVRR63 pKa = 11.84RR64 pKa = 11.84TPHH67 pKa = 6.39RR68 pKa = 11.84VTVPIASSGLRR79 pKa = 11.84PVTTVQYY86 pKa = 11.01DD87 pKa = 3.63PAALSFLLNARR98 pKa = 11.84VDD100 pKa = 3.28WDD102 pKa = 3.98FGNGDD107 pKa = 3.75SANLVINDD115 pKa = 3.19FLFRR119 pKa = 11.84TFAPKK124 pKa = 10.55EE125 pKa = 3.84FDD127 pKa = 3.51FSNSLVPRR135 pKa = 11.84YY136 pKa = 7.01TQAFSAFNAKK146 pKa = 9.28YY147 pKa = 8.72GTMIGEE153 pKa = 4.21GLEE156 pKa = 4.28TIKK159 pKa = 11.09YY160 pKa = 10.14LGLLLRR166 pKa = 11.84RR167 pKa = 11.84LRR169 pKa = 11.84EE170 pKa = 3.85GYY172 pKa = 9.97RR173 pKa = 11.84AVKK176 pKa = 10.45RR177 pKa = 11.84GDD179 pKa = 3.12LRR181 pKa = 11.84ALRR184 pKa = 11.84RR185 pKa = 11.84VIQSYY190 pKa = 11.34HH191 pKa = 5.03NGKK194 pKa = 8.62WKK196 pKa = 9.67PATAGNLWLEE206 pKa = 4.05FRR208 pKa = 11.84YY209 pKa = 10.88GLMPLFYY216 pKa = 10.44DD217 pKa = 3.88IRR219 pKa = 11.84DD220 pKa = 3.92VMLDD224 pKa = 3.02WQNRR228 pKa = 11.84HH229 pKa = 6.37DD230 pKa = 4.62KK231 pKa = 10.31IQRR234 pKa = 11.84LLRR237 pKa = 11.84FSVGHH242 pKa = 6.04GEE244 pKa = 4.23DD245 pKa = 3.78YY246 pKa = 11.19VVEE249 pKa = 4.32FDD251 pKa = 3.66NLYY254 pKa = 10.04PAVAYY259 pKa = 8.42FKK261 pKa = 11.07LKK263 pKa = 11.18GEE265 pKa = 3.9ITLEE269 pKa = 3.51RR270 pKa = 11.84RR271 pKa = 11.84HH272 pKa = 5.59RR273 pKa = 11.84HH274 pKa = 5.18GISYY278 pKa = 10.79ANRR281 pKa = 11.84EE282 pKa = 4.21GYY284 pKa = 10.74AVFDD288 pKa = 4.06NGSLRR293 pKa = 11.84PVSDD297 pKa = 3.0WKK299 pKa = 10.98EE300 pKa = 3.52LATAFINPHH309 pKa = 4.5EE310 pKa = 4.7VAWEE314 pKa = 3.73LTPYY318 pKa = 10.72SFVVDD323 pKa = 3.51WFLNVGDD330 pKa = 4.94ILAQQGQLYY339 pKa = 10.14HH340 pKa = 6.89NIDD343 pKa = 3.27IVDD346 pKa = 3.7GFDD349 pKa = 3.84RR350 pKa = 11.84RR351 pKa = 11.84DD352 pKa = 3.05IRR354 pKa = 11.84LKK356 pKa = 10.49SFTIKK361 pKa = 10.67GEE363 pKa = 4.24RR364 pKa = 11.84NGRR367 pKa = 11.84PVNVSASLSAVDD379 pKa = 4.34LFYY382 pKa = 11.51SRR384 pKa = 11.84LHH386 pKa = 5.8TSNLPFATLDD396 pKa = 3.44LDD398 pKa = 3.97TTFSSFKK405 pKa = 10.41HH406 pKa = 5.3VLDD409 pKa = 4.96SIFLLTQRR417 pKa = 11.84VKK419 pKa = 10.83RR420 pKa = 3.95
Molecular weight: 48.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1471 |
133 |
589 |
367.8 |
41.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.022 ± 0.754 | 1.564 ± 0.628 |
6.662 ± 0.395 | 4.215 ± 0.247 |
4.827 ± 0.793 | 6.662 ± 0.264 |
1.564 ± 0.525 | 5.303 ± 0.653 |
4.487 ± 0.387 | 9.925 ± 0.854 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
0.952 ± 0.116 | 4.895 ± 0.388 |
5.642 ± 0.701 | 2.855 ± 0.629 |
7.206 ± 0.726 | 7.478 ± 0.975 |
5.778 ± 0.661 | 6.526 ± 0.721 |
1.496 ± 0.319 | 3.943 ± 0.163 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
