
Firmicutes bacterium CAG:124
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; environmental samples
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
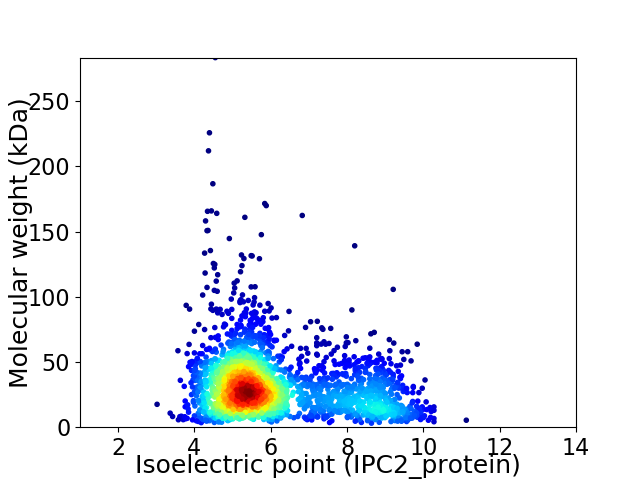
Virtual 2D-PAGE plot for 2364 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5IBY7|R5IBY7_9FIRM Uncharacterized protein OS=Firmicutes bacterium CAG:124 OX=1263002 GN=BN480_00939 PE=4 SV=1
MM1 pKa = 7.61TKK3 pKa = 10.44RR4 pKa = 11.84LLCVCLALVLLFGVLAGCSKK24 pKa = 11.22NEE26 pKa = 3.89AQTADD31 pKa = 3.67DD32 pKa = 3.96TKK34 pKa = 10.57KK35 pKa = 9.13TEE37 pKa = 4.38QSDD40 pKa = 4.31TEE42 pKa = 4.85DD43 pKa = 3.24KK44 pKa = 11.02TSTQFAYY51 pKa = 9.62QAQYY55 pKa = 11.05FDD57 pKa = 4.81LPEE60 pKa = 5.22DD61 pKa = 3.72IQWIGTSCVTGDD73 pKa = 3.63TLYY76 pKa = 9.23FTASVPDD83 pKa = 4.07GGKK86 pKa = 8.21EE87 pKa = 3.98TYY89 pKa = 9.58TDD91 pKa = 3.56EE92 pKa = 4.4NGEE95 pKa = 4.22EE96 pKa = 3.98ISYY99 pKa = 8.41DD100 pKa = 3.47TYY102 pKa = 11.52SEE104 pKa = 4.43VIFRR108 pKa = 11.84FDD110 pKa = 4.38LDD112 pKa = 3.37TGEE115 pKa = 5.42CVQLDD120 pKa = 4.0NYY122 pKa = 9.58VAEE125 pKa = 4.48PVVEE129 pKa = 4.21NPDD132 pKa = 3.52MPDD135 pKa = 3.35GVTMNPDD142 pKa = 2.67GSMQTFNSSTSIQTMAAGADD162 pKa = 3.73GTLWLYY168 pKa = 10.7RR169 pKa = 11.84QTSRR173 pKa = 11.84YY174 pKa = 9.98ADD176 pKa = 4.32DD177 pKa = 3.77GTEE180 pKa = 4.04GDD182 pKa = 4.82SISEE186 pKa = 4.6LIQLDD191 pKa = 3.35AHH193 pKa = 5.81GTLLRR198 pKa = 11.84TITPVSDD205 pKa = 3.71EE206 pKa = 4.32EE207 pKa = 4.5TDD209 pKa = 4.08DD210 pKa = 3.77TDD212 pKa = 2.76SWRR215 pKa = 11.84YY216 pKa = 7.86TYY218 pKa = 10.31IDD220 pKa = 4.96SILSDD225 pKa = 3.7DD226 pKa = 3.48KK227 pKa = 11.63GYY229 pKa = 11.32VYY231 pKa = 10.18TYY233 pKa = 10.4DD234 pKa = 3.62YY235 pKa = 10.54QTVNVYY241 pKa = 10.54GPDD244 pKa = 3.39GSFVFSKK251 pKa = 10.86SGDD254 pKa = 3.61EE255 pKa = 4.64LNGQICQLSASEE267 pKa = 4.33VGMTTSSADD276 pKa = 2.85GKK278 pKa = 10.04MVFKK282 pKa = 10.91QLDD285 pKa = 4.06PEE287 pKa = 4.36TKK289 pKa = 9.93DD290 pKa = 3.02WGKK293 pKa = 8.21EE294 pKa = 4.08TPVSSRR300 pKa = 11.84AWNILPGNDD309 pKa = 2.63VYY311 pKa = 11.54AYY313 pKa = 10.76FFMDD317 pKa = 3.08NGNIFGEE324 pKa = 4.08RR325 pKa = 11.84RR326 pKa = 11.84DD327 pKa = 3.59NGEE330 pKa = 3.8VEE332 pKa = 5.18KK333 pKa = 11.29VVDD336 pKa = 4.11WVACDD341 pKa = 3.35VDD343 pKa = 4.64SNNINSDD350 pKa = 3.16QFGFLSDD357 pKa = 3.5GRR359 pKa = 11.84IVAVTYY365 pKa = 10.09DD366 pKa = 3.74YY367 pKa = 11.82SDD369 pKa = 4.9DD370 pKa = 4.25GPSKK374 pKa = 10.5QQILVLNRR382 pKa = 11.84VDD384 pKa = 3.55AASVTAKK391 pKa = 10.16TEE393 pKa = 3.87LTLACLYY400 pKa = 10.79LDD402 pKa = 3.67YY403 pKa = 11.02NLRR406 pKa = 11.84SQIVKK411 pKa = 9.97FNKK414 pKa = 9.9SNPDD418 pKa = 3.01YY419 pKa = 10.81RR420 pKa = 11.84IVVKK424 pKa = 10.46DD425 pKa = 3.51YY426 pKa = 11.6SEE428 pKa = 4.24YY429 pKa = 10.76ATDD432 pKa = 3.95DD433 pKa = 4.21DD434 pKa = 5.08YY435 pKa = 12.16NAGLTKK441 pKa = 10.79LNTEE445 pKa = 4.64IISGNVPDD453 pKa = 4.15ILVNGTEE460 pKa = 4.29LPIGQYY466 pKa = 9.19AAKK469 pKa = 10.46GLLEE473 pKa = 4.91DD474 pKa = 4.02LWPYY478 pKa = 11.2LDD480 pKa = 5.38ADD482 pKa = 3.91PEE484 pKa = 4.2YY485 pKa = 11.3SRR487 pKa = 11.84DD488 pKa = 3.48KK489 pKa = 11.51LMSQPLNAAQTDD501 pKa = 3.83GKK503 pKa = 10.69LYY505 pKa = 10.44RR506 pKa = 11.84LPIDD510 pKa = 4.13FGVTTAVGLGKK521 pKa = 10.48VVGEE525 pKa = 4.1YY526 pKa = 7.33TTWTLADD533 pKa = 3.71VNDD536 pKa = 4.71ALSKK540 pKa = 10.41LPEE543 pKa = 4.39GATVFNKK550 pKa = 10.43YY551 pKa = 7.23YY552 pKa = 9.27TQAEE556 pKa = 4.17MLQYY560 pKa = 10.05CIAMNAEE567 pKa = 5.11SFMNWQDD574 pKa = 3.55GTCSFDD580 pKa = 3.31TDD582 pKa = 3.31EE583 pKa = 5.29FRR585 pKa = 11.84ALLEE589 pKa = 4.09FVKK592 pKa = 10.2PFPAEE597 pKa = 3.9YY598 pKa = 9.96DD599 pKa = 3.48WQSDD603 pKa = 3.5SDD605 pKa = 4.74DD606 pKa = 4.04YY607 pKa = 11.84EE608 pKa = 5.05SDD610 pKa = 3.5YY611 pKa = 11.64TRR613 pKa = 11.84LKK615 pKa = 10.73NGKK618 pKa = 8.22QLLYY622 pKa = 8.15PTSLSGFSDD631 pKa = 4.29LYY633 pKa = 10.01YY634 pKa = 10.94TFAALNNDD642 pKa = 2.27IRR644 pKa = 11.84FIGFPRR650 pKa = 11.84EE651 pKa = 4.24DD652 pKa = 3.63GSSGNAFNASCTLSISTTCKK672 pKa = 10.34DD673 pKa = 4.24KK674 pKa = 11.32SGAWAFIRR682 pKa = 11.84STLSDD687 pKa = 4.54DD688 pKa = 3.78YY689 pKa = 11.66QEE691 pKa = 5.63SIWNYY696 pKa = 9.42PIVKK700 pKa = 10.02SVFEE704 pKa = 4.45AKK706 pKa = 10.2AQEE709 pKa = 4.19AMTQEE714 pKa = 4.5YY715 pKa = 7.69EE716 pKa = 3.76TDD718 pKa = 3.33ADD720 pKa = 4.08GNQILDD726 pKa = 4.09DD727 pKa = 5.28DD728 pKa = 5.25GNPIPISSGGMSYY741 pKa = 11.28GNEE744 pKa = 3.88PMIEE748 pKa = 4.31LYY750 pKa = 10.74AVTQEE755 pKa = 4.22QYY757 pKa = 11.37DD758 pKa = 3.77AVLALIDD765 pKa = 3.79STTTFVDD772 pKa = 3.28YY773 pKa = 10.76DD774 pKa = 3.85QNVLDD779 pKa = 5.24IISDD783 pKa = 3.5EE784 pKa = 4.1AAGYY788 pKa = 8.73FAGSKK793 pKa = 8.16TVEE796 pKa = 4.25EE797 pKa = 4.16ASKK800 pKa = 10.69LIQSRR805 pKa = 11.84VSLYY809 pKa = 10.27IQEE812 pKa = 4.22QKK814 pKa = 11.15
MM1 pKa = 7.61TKK3 pKa = 10.44RR4 pKa = 11.84LLCVCLALVLLFGVLAGCSKK24 pKa = 11.22NEE26 pKa = 3.89AQTADD31 pKa = 3.67DD32 pKa = 3.96TKK34 pKa = 10.57KK35 pKa = 9.13TEE37 pKa = 4.38QSDD40 pKa = 4.31TEE42 pKa = 4.85DD43 pKa = 3.24KK44 pKa = 11.02TSTQFAYY51 pKa = 9.62QAQYY55 pKa = 11.05FDD57 pKa = 4.81LPEE60 pKa = 5.22DD61 pKa = 3.72IQWIGTSCVTGDD73 pKa = 3.63TLYY76 pKa = 9.23FTASVPDD83 pKa = 4.07GGKK86 pKa = 8.21EE87 pKa = 3.98TYY89 pKa = 9.58TDD91 pKa = 3.56EE92 pKa = 4.4NGEE95 pKa = 4.22EE96 pKa = 3.98ISYY99 pKa = 8.41DD100 pKa = 3.47TYY102 pKa = 11.52SEE104 pKa = 4.43VIFRR108 pKa = 11.84FDD110 pKa = 4.38LDD112 pKa = 3.37TGEE115 pKa = 5.42CVQLDD120 pKa = 4.0NYY122 pKa = 9.58VAEE125 pKa = 4.48PVVEE129 pKa = 4.21NPDD132 pKa = 3.52MPDD135 pKa = 3.35GVTMNPDD142 pKa = 2.67GSMQTFNSSTSIQTMAAGADD162 pKa = 3.73GTLWLYY168 pKa = 10.7RR169 pKa = 11.84QTSRR173 pKa = 11.84YY174 pKa = 9.98ADD176 pKa = 4.32DD177 pKa = 3.77GTEE180 pKa = 4.04GDD182 pKa = 4.82SISEE186 pKa = 4.6LIQLDD191 pKa = 3.35AHH193 pKa = 5.81GTLLRR198 pKa = 11.84TITPVSDD205 pKa = 3.71EE206 pKa = 4.32EE207 pKa = 4.5TDD209 pKa = 4.08DD210 pKa = 3.77TDD212 pKa = 2.76SWRR215 pKa = 11.84YY216 pKa = 7.86TYY218 pKa = 10.31IDD220 pKa = 4.96SILSDD225 pKa = 3.7DD226 pKa = 3.48KK227 pKa = 11.63GYY229 pKa = 11.32VYY231 pKa = 10.18TYY233 pKa = 10.4DD234 pKa = 3.62YY235 pKa = 10.54QTVNVYY241 pKa = 10.54GPDD244 pKa = 3.39GSFVFSKK251 pKa = 10.86SGDD254 pKa = 3.61EE255 pKa = 4.64LNGQICQLSASEE267 pKa = 4.33VGMTTSSADD276 pKa = 2.85GKK278 pKa = 10.04MVFKK282 pKa = 10.91QLDD285 pKa = 4.06PEE287 pKa = 4.36TKK289 pKa = 9.93DD290 pKa = 3.02WGKK293 pKa = 8.21EE294 pKa = 4.08TPVSSRR300 pKa = 11.84AWNILPGNDD309 pKa = 2.63VYY311 pKa = 11.54AYY313 pKa = 10.76FFMDD317 pKa = 3.08NGNIFGEE324 pKa = 4.08RR325 pKa = 11.84RR326 pKa = 11.84DD327 pKa = 3.59NGEE330 pKa = 3.8VEE332 pKa = 5.18KK333 pKa = 11.29VVDD336 pKa = 4.11WVACDD341 pKa = 3.35VDD343 pKa = 4.64SNNINSDD350 pKa = 3.16QFGFLSDD357 pKa = 3.5GRR359 pKa = 11.84IVAVTYY365 pKa = 10.09DD366 pKa = 3.74YY367 pKa = 11.82SDD369 pKa = 4.9DD370 pKa = 4.25GPSKK374 pKa = 10.5QQILVLNRR382 pKa = 11.84VDD384 pKa = 3.55AASVTAKK391 pKa = 10.16TEE393 pKa = 3.87LTLACLYY400 pKa = 10.79LDD402 pKa = 3.67YY403 pKa = 11.02NLRR406 pKa = 11.84SQIVKK411 pKa = 9.97FNKK414 pKa = 9.9SNPDD418 pKa = 3.01YY419 pKa = 10.81RR420 pKa = 11.84IVVKK424 pKa = 10.46DD425 pKa = 3.51YY426 pKa = 11.6SEE428 pKa = 4.24YY429 pKa = 10.76ATDD432 pKa = 3.95DD433 pKa = 4.21DD434 pKa = 5.08YY435 pKa = 12.16NAGLTKK441 pKa = 10.79LNTEE445 pKa = 4.64IISGNVPDD453 pKa = 4.15ILVNGTEE460 pKa = 4.29LPIGQYY466 pKa = 9.19AAKK469 pKa = 10.46GLLEE473 pKa = 4.91DD474 pKa = 4.02LWPYY478 pKa = 11.2LDD480 pKa = 5.38ADD482 pKa = 3.91PEE484 pKa = 4.2YY485 pKa = 11.3SRR487 pKa = 11.84DD488 pKa = 3.48KK489 pKa = 11.51LMSQPLNAAQTDD501 pKa = 3.83GKK503 pKa = 10.69LYY505 pKa = 10.44RR506 pKa = 11.84LPIDD510 pKa = 4.13FGVTTAVGLGKK521 pKa = 10.48VVGEE525 pKa = 4.1YY526 pKa = 7.33TTWTLADD533 pKa = 3.71VNDD536 pKa = 4.71ALSKK540 pKa = 10.41LPEE543 pKa = 4.39GATVFNKK550 pKa = 10.43YY551 pKa = 7.23YY552 pKa = 9.27TQAEE556 pKa = 4.17MLQYY560 pKa = 10.05CIAMNAEE567 pKa = 5.11SFMNWQDD574 pKa = 3.55GTCSFDD580 pKa = 3.31TDD582 pKa = 3.31EE583 pKa = 5.29FRR585 pKa = 11.84ALLEE589 pKa = 4.09FVKK592 pKa = 10.2PFPAEE597 pKa = 3.9YY598 pKa = 9.96DD599 pKa = 3.48WQSDD603 pKa = 3.5SDD605 pKa = 4.74DD606 pKa = 4.04YY607 pKa = 11.84EE608 pKa = 5.05SDD610 pKa = 3.5YY611 pKa = 11.64TRR613 pKa = 11.84LKK615 pKa = 10.73NGKK618 pKa = 8.22QLLYY622 pKa = 8.15PTSLSGFSDD631 pKa = 4.29LYY633 pKa = 10.01YY634 pKa = 10.94TFAALNNDD642 pKa = 2.27IRR644 pKa = 11.84FIGFPRR650 pKa = 11.84EE651 pKa = 4.24DD652 pKa = 3.63GSSGNAFNASCTLSISTTCKK672 pKa = 10.34DD673 pKa = 4.24KK674 pKa = 11.32SGAWAFIRR682 pKa = 11.84STLSDD687 pKa = 4.54DD688 pKa = 3.78YY689 pKa = 11.66QEE691 pKa = 5.63SIWNYY696 pKa = 9.42PIVKK700 pKa = 10.02SVFEE704 pKa = 4.45AKK706 pKa = 10.2AQEE709 pKa = 4.19AMTQEE714 pKa = 4.5YY715 pKa = 7.69EE716 pKa = 3.76TDD718 pKa = 3.33ADD720 pKa = 4.08GNQILDD726 pKa = 4.09DD727 pKa = 5.28DD728 pKa = 5.25GNPIPISSGGMSYY741 pKa = 11.28GNEE744 pKa = 3.88PMIEE748 pKa = 4.31LYY750 pKa = 10.74AVTQEE755 pKa = 4.22QYY757 pKa = 11.37DD758 pKa = 3.77AVLALIDD765 pKa = 3.79STTTFVDD772 pKa = 3.28YY773 pKa = 10.76DD774 pKa = 3.85QNVLDD779 pKa = 5.24IISDD783 pKa = 3.5EE784 pKa = 4.1AAGYY788 pKa = 8.73FAGSKK793 pKa = 8.16TVEE796 pKa = 4.25EE797 pKa = 4.16ASKK800 pKa = 10.69LIQSRR805 pKa = 11.84VSLYY809 pKa = 10.27IQEE812 pKa = 4.22QKK814 pKa = 11.15
Molecular weight: 90.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5IIM2|R5IIM2_9FIRM Filamentation protein Fic1 OS=Firmicutes bacterium CAG:124 OX=1263002 GN=BN480_00069 PE=4 SV=1
MM1 pKa = 7.09ATVRR5 pKa = 11.84TYY7 pKa = 10.02QPRR10 pKa = 11.84KK11 pKa = 7.15RR12 pKa = 11.84HH13 pKa = 5.32RR14 pKa = 11.84SKK16 pKa = 10.5VHH18 pKa = 5.66GFRR21 pKa = 11.84KK22 pKa = 10.08RR23 pKa = 11.84MATANGRR30 pKa = 11.84KK31 pKa = 8.93VLARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84AKK39 pKa = 10.49GRR41 pKa = 11.84AKK43 pKa = 10.69LSAA46 pKa = 3.92
MM1 pKa = 7.09ATVRR5 pKa = 11.84TYY7 pKa = 10.02QPRR10 pKa = 11.84KK11 pKa = 7.15RR12 pKa = 11.84HH13 pKa = 5.32RR14 pKa = 11.84SKK16 pKa = 10.5VHH18 pKa = 5.66GFRR21 pKa = 11.84KK22 pKa = 10.08RR23 pKa = 11.84MATANGRR30 pKa = 11.84KK31 pKa = 8.93VLARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84AKK39 pKa = 10.49GRR41 pKa = 11.84AKK43 pKa = 10.69LSAA46 pKa = 3.92
Molecular weight: 5.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
714807 |
29 |
2678 |
302.4 |
33.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.31 ± 0.057 | 1.855 ± 0.027 |
5.61 ± 0.045 | 6.547 ± 0.047 |
3.784 ± 0.035 | 7.59 ± 0.047 |
1.801 ± 0.023 | 5.643 ± 0.049 |
4.79 ± 0.049 | 9.96 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.726 ± 0.026 | 3.252 ± 0.031 |
4.122 ± 0.035 | 3.643 ± 0.034 |
5.544 ± 0.054 | 5.54 ± 0.043 |
5.808 ± 0.056 | 6.848 ± 0.036 |
1.016 ± 0.021 | 3.61 ± 0.042 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
