
Thioclava sp. SK-1
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Thioclava; unclassified Thioclava
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
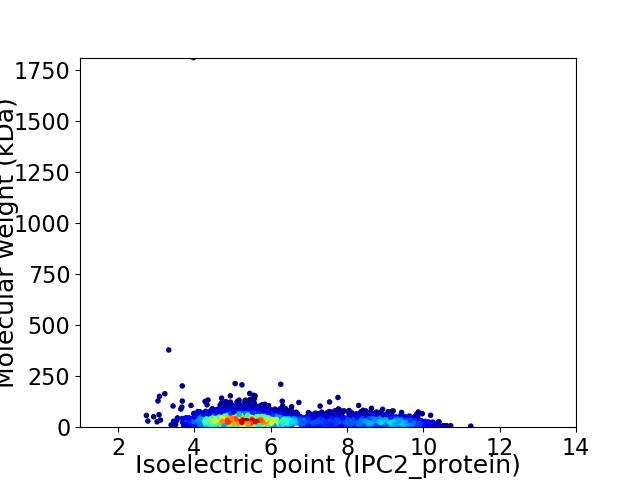
Virtual 2D-PAGE plot for 3819 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1C2HNN6|A0A1C2HNN6_9RHOB Peptidase_M48 domain-containing protein OS=Thioclava sp. SK-1 OX=1889770 GN=BFP70_09865 PE=3 SV=1
MM1 pKa = 7.74AILWGLGGPAAHH13 pKa = 7.12AGDD16 pKa = 4.51FQIDD20 pKa = 3.34WEE22 pKa = 4.37AFDD25 pKa = 3.98WPAGVTGPLVRR36 pKa = 11.84TLRR39 pKa = 11.84DD40 pKa = 3.36QNGFEE45 pKa = 4.08VDD47 pKa = 3.45VTVTHH52 pKa = 6.95TGPFAGYY59 pKa = 8.93PDD61 pKa = 4.31GAGGTIPTPDD71 pKa = 3.92DD72 pKa = 3.25VTIFGGSTEE81 pKa = 4.12SLILVVDD88 pKa = 4.58APQNQGAIGDD98 pKa = 3.96SRR100 pKa = 11.84TISTVSASSGGIAILVDD117 pKa = 4.05DD118 pKa = 4.51LTIDD122 pKa = 3.61VFDD125 pKa = 5.55IDD127 pKa = 4.01STDD130 pKa = 3.3NNAVSDD136 pKa = 3.84RR137 pKa = 11.84CDD139 pKa = 3.3FLTVFGDD146 pKa = 3.47NGNPTLSTLSLTPSVVVGPGFGSGLTGPITANQAQCIYY184 pKa = 10.77FEE186 pKa = 5.12GPTGSPTSPNDD197 pKa = 3.17DD198 pKa = 3.76TGSVRR203 pKa = 11.84ATFPDD208 pKa = 3.39ATSSVTFWYY217 pKa = 10.15DD218 pKa = 3.06EE219 pKa = 4.41SIQNVRR225 pKa = 11.84NYY227 pKa = 11.07NVFTTYY233 pKa = 11.2DD234 pKa = 3.23PGARR238 pKa = 11.84GLGMFGNANFTVDD251 pKa = 3.64QSISLARR258 pKa = 11.84SVTPISGLQGDD269 pKa = 4.58TVTYY273 pKa = 10.48SYY275 pKa = 10.76TVTNNGEE282 pKa = 3.99LPFNIGQDD290 pKa = 3.65VVIEE294 pKa = 4.44DD295 pKa = 4.56DD296 pKa = 3.98LLGSVTCPAITAPIAPGGTVTCTAPYY322 pKa = 9.01TISALDD328 pKa = 3.65VLTGTVNSTATAGIGAIGQAFSARR352 pKa = 11.84LQSNSQSLSLVTSVLSGDD370 pKa = 4.31SGAQNCTPQSVFAHH384 pKa = 5.84TRR386 pKa = 11.84SQLAGPGSAAALTTSDD402 pKa = 3.72IFLFDD407 pKa = 5.34DD408 pKa = 3.37VTEE411 pKa = 4.53DD412 pKa = 3.58VNGNAIDD419 pKa = 4.08VVFQLDD425 pKa = 3.73QISNASAVKK434 pKa = 10.8LEE436 pKa = 4.37GSLQALMTPSNNGYY450 pKa = 7.48VTYY453 pKa = 10.66RR454 pKa = 11.84LRR456 pKa = 11.84LVQDD460 pKa = 4.11GTATPANPQGIAIEE474 pKa = 3.93QSRR477 pKa = 11.84INGVIVQQTDD487 pKa = 2.56VDD489 pKa = 4.23SRR491 pKa = 11.84GSSDD495 pKa = 3.83DD496 pKa = 3.48SSDD499 pKa = 3.39VVGPVSSPRR508 pKa = 11.84AISYY512 pKa = 10.77LNTAPLVAFPAPGQAIAMDD531 pKa = 4.42PAKK534 pKa = 10.75QGDD537 pKa = 3.72PSNWIDD543 pKa = 4.37EE544 pKa = 4.47PNEE547 pKa = 4.21TNFDD551 pKa = 3.38NYY553 pKa = 9.98ATYY556 pKa = 10.19EE557 pKa = 4.0YY558 pKa = 9.64DD559 pKa = 3.48TFIEE563 pKa = 4.72GAFIHH568 pKa = 6.84GFTGSSTNPATRR580 pKa = 11.84GSGILLCGIVNSSADD595 pKa = 3.56VIARR599 pKa = 11.84DD600 pKa = 4.07DD601 pKa = 5.03DD602 pKa = 4.3YY603 pKa = 11.51TASPLNSLLGGTAGEE618 pKa = 4.22VLVNDD623 pKa = 5.5TINALPALFPTATLEE638 pKa = 4.2VLTPAVPQNDD648 pKa = 3.96GDD650 pKa = 4.1PVPVLEE656 pKa = 4.56TSGVDD661 pKa = 2.98AGRR664 pKa = 11.84VIVPPGVPAGTYY676 pKa = 8.92TIDD679 pKa = 3.8YY680 pKa = 8.86RR681 pKa = 11.84LCDD684 pKa = 3.47SLTPGDD690 pKa = 3.98CDD692 pKa = 3.28RR693 pKa = 11.84AKK695 pKa = 10.03VTLAVFDD702 pKa = 4.1GLGLDD707 pKa = 4.32FGDD710 pKa = 4.51APVTYY715 pKa = 10.06LVAAHH720 pKa = 7.05AVRR723 pKa = 11.84PSPTIYY729 pKa = 10.22LGSVAPDD736 pKa = 3.29IEE738 pKa = 4.52IVAQSDD744 pKa = 3.41ATATADD750 pKa = 4.96DD751 pKa = 5.76LIDD754 pKa = 4.88NDD756 pKa = 5.19DD757 pKa = 3.63EE758 pKa = 4.55DD759 pKa = 6.91AILFPVMTQGAISTLNIPVTGAGVLQAWIDD789 pKa = 3.85FNGDD793 pKa = 3.2GVFEE797 pKa = 4.23EE798 pKa = 4.64TLGEE802 pKa = 4.43RR803 pKa = 11.84IASDD807 pKa = 3.94LSDD810 pKa = 4.79DD811 pKa = 3.69GTGFDD816 pKa = 3.94TVAGDD821 pKa = 3.69GVIQVDD827 pKa = 3.87VAVPTDD833 pKa = 3.2ATTATTYY840 pKa = 11.35ARR842 pKa = 11.84FRR844 pKa = 11.84YY845 pKa = 9.66ASTPGLPAAGFALDD859 pKa = 5.22GEE861 pKa = 4.69VEE863 pKa = 4.54DD864 pKa = 4.27YY865 pKa = 11.55SLVIVAADD873 pKa = 3.44LVDD876 pKa = 5.01RR877 pKa = 11.84GDD879 pKa = 3.89APASYY884 pKa = 10.51GDD886 pKa = 3.63PRR888 pKa = 11.84HH889 pKa = 6.06IVVPSIYY896 pKa = 10.26LGAGLPDD903 pKa = 4.2TEE905 pKa = 4.4WVPQNSVTADD915 pKa = 3.54ADD917 pKa = 3.91DD918 pKa = 4.44LAGSDD923 pKa = 4.79DD924 pKa = 4.01EE925 pKa = 5.39DD926 pKa = 6.34AIALFPVLEE935 pKa = 4.95AGTTQNLTVQTHH947 pKa = 5.04EE948 pKa = 4.25TLSLQLALGLPVLSGITNLQLWVDD972 pKa = 3.78WDD974 pKa = 3.56QNGRR978 pKa = 11.84FEE980 pKa = 4.23TSEE983 pKa = 3.9QVAVNYY989 pKa = 10.09RR990 pKa = 11.84DD991 pKa = 3.57GGAGDD996 pKa = 3.58TDD998 pKa = 3.83GVFNNQISLEE1008 pKa = 4.1IPVPGDD1014 pKa = 2.73IGNGISYY1021 pKa = 10.9ARR1023 pKa = 11.84VRR1025 pKa = 11.84WSTTSIVGLEE1035 pKa = 4.18PFDD1038 pKa = 4.1GLNLDD1043 pKa = 4.81GEE1045 pKa = 4.67VEE1047 pKa = 4.5DD1048 pKa = 4.83YY1049 pKa = 11.25LVTLSNPNAPLVCDD1063 pKa = 3.13TGFYY1067 pKa = 9.81MIYY1070 pKa = 10.34EE1071 pKa = 4.42SGAQPILEE1079 pKa = 4.43KK1080 pKa = 10.97LQISGSAGSYY1090 pKa = 7.34STNGTTYY1097 pKa = 9.62PANFSGTYY1105 pKa = 9.18TMSGWGWNEE1114 pKa = 3.16VDD1116 pKa = 3.39NYY1118 pKa = 10.4IYY1120 pKa = 10.82GIFRR1124 pKa = 11.84NTFQLHH1130 pKa = 5.19RR1131 pKa = 11.84VTYY1134 pKa = 9.13SGAVQLVSDD1143 pKa = 4.29MSALGLEE1150 pKa = 4.53PVGTTLEE1157 pKa = 4.0ILPNGVLIYY1166 pKa = 10.58NSQATTGRR1174 pKa = 11.84YY1175 pKa = 7.83QLVDD1179 pKa = 3.25ISDD1182 pKa = 3.98PSAPTNLGVLEE1193 pKa = 5.07AGSSAPNGLDD1203 pKa = 2.93MAYY1206 pKa = 10.52NPRR1209 pKa = 11.84DD1210 pKa = 3.18GLLYY1214 pKa = 10.06MVANGNDD1221 pKa = 3.43IYY1223 pKa = 11.53ALDD1226 pKa = 4.28PLGGVAGATSTVLVADD1242 pKa = 4.14NVTLPAGTGSYY1253 pKa = 10.51QLDD1256 pKa = 4.1SVWMDD1261 pKa = 3.19EE1262 pKa = 3.79NGYY1265 pKa = 10.33FYY1267 pKa = 10.66AYY1269 pKa = 10.63DD1270 pKa = 3.62NVSQQIFALEE1280 pKa = 3.96LGEE1283 pKa = 5.26AGDD1286 pKa = 4.52RR1287 pKa = 11.84PASFQFFKK1295 pKa = 11.26VDD1297 pKa = 3.74GTGSAQIGNDD1307 pKa = 3.36GASCRR1312 pKa = 11.84KK1313 pKa = 9.27GSFYY1317 pKa = 11.08ASTVFAEE1324 pKa = 4.73GTISGRR1330 pKa = 11.84LYY1332 pKa = 10.58HH1333 pKa = 7.35DD1334 pKa = 4.7SNASGAYY1341 pKa = 9.61DD1342 pKa = 3.39VGEE1345 pKa = 4.5SGLPADD1351 pKa = 4.62ISMALYY1357 pKa = 10.23RR1358 pKa = 11.84DD1359 pKa = 3.46NATPADD1365 pKa = 4.18LTDD1368 pKa = 5.78DD1369 pKa = 3.61ILVTTTQTDD1378 pKa = 3.18GDD1380 pKa = 3.81GRR1382 pKa = 11.84YY1383 pKa = 9.46VFAGVDD1389 pKa = 3.19ATVTYY1394 pKa = 10.35RR1395 pKa = 11.84IEE1397 pKa = 4.09VNEE1400 pKa = 4.65ADD1402 pKa = 4.18PEE1404 pKa = 4.23IPANRR1409 pKa = 11.84TISTPNPRR1417 pKa = 11.84IGVTVMTGAEE1427 pKa = 4.15TSDD1430 pKa = 3.13QDD1432 pKa = 4.14FGFEE1436 pKa = 3.9ATALQADD1443 pKa = 4.54LSLNKK1448 pKa = 10.14AAFDD1452 pKa = 3.67TSGAALSSAQDD1463 pKa = 3.44GAQIDD1468 pKa = 4.03FVLTVTNDD1476 pKa = 3.44GPATATDD1483 pKa = 3.36VRR1485 pKa = 11.84VRR1487 pKa = 11.84DD1488 pKa = 4.61LIPDD1492 pKa = 4.18GFTYY1496 pKa = 10.7VSNDD1500 pKa = 2.83AAAQAMGYY1508 pKa = 10.06DD1509 pKa = 4.0PSSGIWTLTDD1519 pKa = 3.87LPDD1522 pKa = 3.88GTSATLTLRR1531 pKa = 11.84VVMKK1535 pKa = 10.1ATGTHH1540 pKa = 5.99TNTAEE1545 pKa = 4.16IVASAIPDD1553 pKa = 4.14PDD1555 pKa = 3.69SDD1557 pKa = 4.16PAVGTLRR1564 pKa = 11.84DD1565 pKa = 4.19DD1566 pKa = 4.17LSDD1569 pKa = 4.93GIVDD1573 pKa = 4.55DD1574 pKa = 5.49DD1575 pKa = 4.11EE1576 pKa = 7.04ASATVSFIGMGPTLSGTVFRR1596 pKa = 11.84DD1597 pKa = 3.0NGAAGDD1603 pKa = 3.73TAYY1606 pKa = 10.77DD1607 pKa = 3.95GLQGAQEE1614 pKa = 4.16PGIPDD1619 pKa = 3.35ATVTVADD1626 pKa = 3.78TVGTVLGQPVVAADD1640 pKa = 4.13GTWSLTLPEE1649 pKa = 4.85GFNSAVTVTLAPTSDD1664 pKa = 3.39GQVVSEE1670 pKa = 4.99SPGTLPGLVNTQARR1684 pKa = 11.84DD1685 pKa = 3.29GSFGFTPVSGGTYY1698 pKa = 9.86SNLNFGVIPSARR1710 pKa = 11.84LRR1712 pKa = 11.84NSQQAAMRR1720 pKa = 11.84SGQVITLRR1728 pKa = 11.84HH1729 pKa = 6.5EE1730 pKa = 4.29YY1731 pKa = 8.85TAQAPGNVTFEE1742 pKa = 4.33IDD1744 pKa = 3.28VHH1746 pKa = 5.35QATMPGLFSTGIFLDD1761 pKa = 4.29PACNGTPITAVNGAQPITADD1781 pKa = 3.3TRR1783 pKa = 11.84ICLVIRR1789 pKa = 11.84VTASSAASPGASIAFDD1805 pKa = 4.86LIADD1809 pKa = 4.13TTYY1812 pKa = 11.4GATGLSAQVRR1822 pKa = 11.84NTDD1825 pKa = 3.2VVRR1828 pKa = 11.84VDD1830 pKa = 3.53SSQGQLTLRR1839 pKa = 11.84KK1840 pKa = 7.12TVRR1843 pKa = 11.84NITQGTPEE1851 pKa = 4.27GVSNGAALGDD1861 pKa = 3.56VLEE1864 pKa = 4.03YY1865 pKa = 10.38RR1866 pKa = 11.84IYY1868 pKa = 10.99LDD1870 pKa = 4.36NPGTLPATQIVIHH1883 pKa = 7.11DD1884 pKa = 4.06RR1885 pKa = 11.84TPPYY1889 pKa = 7.57TTLAAPIPSSTAIGTDD1905 pKa = 3.87VICTLAVPSPNNAGYY1920 pKa = 10.53AGPLRR1925 pKa = 11.84WEE1927 pKa = 4.7CAGTHH1932 pKa = 6.22PPGATGSVTFQVQIAPP1948 pKa = 3.74
MM1 pKa = 7.74AILWGLGGPAAHH13 pKa = 7.12AGDD16 pKa = 4.51FQIDD20 pKa = 3.34WEE22 pKa = 4.37AFDD25 pKa = 3.98WPAGVTGPLVRR36 pKa = 11.84TLRR39 pKa = 11.84DD40 pKa = 3.36QNGFEE45 pKa = 4.08VDD47 pKa = 3.45VTVTHH52 pKa = 6.95TGPFAGYY59 pKa = 8.93PDD61 pKa = 4.31GAGGTIPTPDD71 pKa = 3.92DD72 pKa = 3.25VTIFGGSTEE81 pKa = 4.12SLILVVDD88 pKa = 4.58APQNQGAIGDD98 pKa = 3.96SRR100 pKa = 11.84TISTVSASSGGIAILVDD117 pKa = 4.05DD118 pKa = 4.51LTIDD122 pKa = 3.61VFDD125 pKa = 5.55IDD127 pKa = 4.01STDD130 pKa = 3.3NNAVSDD136 pKa = 3.84RR137 pKa = 11.84CDD139 pKa = 3.3FLTVFGDD146 pKa = 3.47NGNPTLSTLSLTPSVVVGPGFGSGLTGPITANQAQCIYY184 pKa = 10.77FEE186 pKa = 5.12GPTGSPTSPNDD197 pKa = 3.17DD198 pKa = 3.76TGSVRR203 pKa = 11.84ATFPDD208 pKa = 3.39ATSSVTFWYY217 pKa = 10.15DD218 pKa = 3.06EE219 pKa = 4.41SIQNVRR225 pKa = 11.84NYY227 pKa = 11.07NVFTTYY233 pKa = 11.2DD234 pKa = 3.23PGARR238 pKa = 11.84GLGMFGNANFTVDD251 pKa = 3.64QSISLARR258 pKa = 11.84SVTPISGLQGDD269 pKa = 4.58TVTYY273 pKa = 10.48SYY275 pKa = 10.76TVTNNGEE282 pKa = 3.99LPFNIGQDD290 pKa = 3.65VVIEE294 pKa = 4.44DD295 pKa = 4.56DD296 pKa = 3.98LLGSVTCPAITAPIAPGGTVTCTAPYY322 pKa = 9.01TISALDD328 pKa = 3.65VLTGTVNSTATAGIGAIGQAFSARR352 pKa = 11.84LQSNSQSLSLVTSVLSGDD370 pKa = 4.31SGAQNCTPQSVFAHH384 pKa = 5.84TRR386 pKa = 11.84SQLAGPGSAAALTTSDD402 pKa = 3.72IFLFDD407 pKa = 5.34DD408 pKa = 3.37VTEE411 pKa = 4.53DD412 pKa = 3.58VNGNAIDD419 pKa = 4.08VVFQLDD425 pKa = 3.73QISNASAVKK434 pKa = 10.8LEE436 pKa = 4.37GSLQALMTPSNNGYY450 pKa = 7.48VTYY453 pKa = 10.66RR454 pKa = 11.84LRR456 pKa = 11.84LVQDD460 pKa = 4.11GTATPANPQGIAIEE474 pKa = 3.93QSRR477 pKa = 11.84INGVIVQQTDD487 pKa = 2.56VDD489 pKa = 4.23SRR491 pKa = 11.84GSSDD495 pKa = 3.83DD496 pKa = 3.48SSDD499 pKa = 3.39VVGPVSSPRR508 pKa = 11.84AISYY512 pKa = 10.77LNTAPLVAFPAPGQAIAMDD531 pKa = 4.42PAKK534 pKa = 10.75QGDD537 pKa = 3.72PSNWIDD543 pKa = 4.37EE544 pKa = 4.47PNEE547 pKa = 4.21TNFDD551 pKa = 3.38NYY553 pKa = 9.98ATYY556 pKa = 10.19EE557 pKa = 4.0YY558 pKa = 9.64DD559 pKa = 3.48TFIEE563 pKa = 4.72GAFIHH568 pKa = 6.84GFTGSSTNPATRR580 pKa = 11.84GSGILLCGIVNSSADD595 pKa = 3.56VIARR599 pKa = 11.84DD600 pKa = 4.07DD601 pKa = 5.03DD602 pKa = 4.3YY603 pKa = 11.51TASPLNSLLGGTAGEE618 pKa = 4.22VLVNDD623 pKa = 5.5TINALPALFPTATLEE638 pKa = 4.2VLTPAVPQNDD648 pKa = 3.96GDD650 pKa = 4.1PVPVLEE656 pKa = 4.56TSGVDD661 pKa = 2.98AGRR664 pKa = 11.84VIVPPGVPAGTYY676 pKa = 8.92TIDD679 pKa = 3.8YY680 pKa = 8.86RR681 pKa = 11.84LCDD684 pKa = 3.47SLTPGDD690 pKa = 3.98CDD692 pKa = 3.28RR693 pKa = 11.84AKK695 pKa = 10.03VTLAVFDD702 pKa = 4.1GLGLDD707 pKa = 4.32FGDD710 pKa = 4.51APVTYY715 pKa = 10.06LVAAHH720 pKa = 7.05AVRR723 pKa = 11.84PSPTIYY729 pKa = 10.22LGSVAPDD736 pKa = 3.29IEE738 pKa = 4.52IVAQSDD744 pKa = 3.41ATATADD750 pKa = 4.96DD751 pKa = 5.76LIDD754 pKa = 4.88NDD756 pKa = 5.19DD757 pKa = 3.63EE758 pKa = 4.55DD759 pKa = 6.91AILFPVMTQGAISTLNIPVTGAGVLQAWIDD789 pKa = 3.85FNGDD793 pKa = 3.2GVFEE797 pKa = 4.23EE798 pKa = 4.64TLGEE802 pKa = 4.43RR803 pKa = 11.84IASDD807 pKa = 3.94LSDD810 pKa = 4.79DD811 pKa = 3.69GTGFDD816 pKa = 3.94TVAGDD821 pKa = 3.69GVIQVDD827 pKa = 3.87VAVPTDD833 pKa = 3.2ATTATTYY840 pKa = 11.35ARR842 pKa = 11.84FRR844 pKa = 11.84YY845 pKa = 9.66ASTPGLPAAGFALDD859 pKa = 5.22GEE861 pKa = 4.69VEE863 pKa = 4.54DD864 pKa = 4.27YY865 pKa = 11.55SLVIVAADD873 pKa = 3.44LVDD876 pKa = 5.01RR877 pKa = 11.84GDD879 pKa = 3.89APASYY884 pKa = 10.51GDD886 pKa = 3.63PRR888 pKa = 11.84HH889 pKa = 6.06IVVPSIYY896 pKa = 10.26LGAGLPDD903 pKa = 4.2TEE905 pKa = 4.4WVPQNSVTADD915 pKa = 3.54ADD917 pKa = 3.91DD918 pKa = 4.44LAGSDD923 pKa = 4.79DD924 pKa = 4.01EE925 pKa = 5.39DD926 pKa = 6.34AIALFPVLEE935 pKa = 4.95AGTTQNLTVQTHH947 pKa = 5.04EE948 pKa = 4.25TLSLQLALGLPVLSGITNLQLWVDD972 pKa = 3.78WDD974 pKa = 3.56QNGRR978 pKa = 11.84FEE980 pKa = 4.23TSEE983 pKa = 3.9QVAVNYY989 pKa = 10.09RR990 pKa = 11.84DD991 pKa = 3.57GGAGDD996 pKa = 3.58TDD998 pKa = 3.83GVFNNQISLEE1008 pKa = 4.1IPVPGDD1014 pKa = 2.73IGNGISYY1021 pKa = 10.9ARR1023 pKa = 11.84VRR1025 pKa = 11.84WSTTSIVGLEE1035 pKa = 4.18PFDD1038 pKa = 4.1GLNLDD1043 pKa = 4.81GEE1045 pKa = 4.67VEE1047 pKa = 4.5DD1048 pKa = 4.83YY1049 pKa = 11.25LVTLSNPNAPLVCDD1063 pKa = 3.13TGFYY1067 pKa = 9.81MIYY1070 pKa = 10.34EE1071 pKa = 4.42SGAQPILEE1079 pKa = 4.43KK1080 pKa = 10.97LQISGSAGSYY1090 pKa = 7.34STNGTTYY1097 pKa = 9.62PANFSGTYY1105 pKa = 9.18TMSGWGWNEE1114 pKa = 3.16VDD1116 pKa = 3.39NYY1118 pKa = 10.4IYY1120 pKa = 10.82GIFRR1124 pKa = 11.84NTFQLHH1130 pKa = 5.19RR1131 pKa = 11.84VTYY1134 pKa = 9.13SGAVQLVSDD1143 pKa = 4.29MSALGLEE1150 pKa = 4.53PVGTTLEE1157 pKa = 4.0ILPNGVLIYY1166 pKa = 10.58NSQATTGRR1174 pKa = 11.84YY1175 pKa = 7.83QLVDD1179 pKa = 3.25ISDD1182 pKa = 3.98PSAPTNLGVLEE1193 pKa = 5.07AGSSAPNGLDD1203 pKa = 2.93MAYY1206 pKa = 10.52NPRR1209 pKa = 11.84DD1210 pKa = 3.18GLLYY1214 pKa = 10.06MVANGNDD1221 pKa = 3.43IYY1223 pKa = 11.53ALDD1226 pKa = 4.28PLGGVAGATSTVLVADD1242 pKa = 4.14NVTLPAGTGSYY1253 pKa = 10.51QLDD1256 pKa = 4.1SVWMDD1261 pKa = 3.19EE1262 pKa = 3.79NGYY1265 pKa = 10.33FYY1267 pKa = 10.66AYY1269 pKa = 10.63DD1270 pKa = 3.62NVSQQIFALEE1280 pKa = 3.96LGEE1283 pKa = 5.26AGDD1286 pKa = 4.52RR1287 pKa = 11.84PASFQFFKK1295 pKa = 11.26VDD1297 pKa = 3.74GTGSAQIGNDD1307 pKa = 3.36GASCRR1312 pKa = 11.84KK1313 pKa = 9.27GSFYY1317 pKa = 11.08ASTVFAEE1324 pKa = 4.73GTISGRR1330 pKa = 11.84LYY1332 pKa = 10.58HH1333 pKa = 7.35DD1334 pKa = 4.7SNASGAYY1341 pKa = 9.61DD1342 pKa = 3.39VGEE1345 pKa = 4.5SGLPADD1351 pKa = 4.62ISMALYY1357 pKa = 10.23RR1358 pKa = 11.84DD1359 pKa = 3.46NATPADD1365 pKa = 4.18LTDD1368 pKa = 5.78DD1369 pKa = 3.61ILVTTTQTDD1378 pKa = 3.18GDD1380 pKa = 3.81GRR1382 pKa = 11.84YY1383 pKa = 9.46VFAGVDD1389 pKa = 3.19ATVTYY1394 pKa = 10.35RR1395 pKa = 11.84IEE1397 pKa = 4.09VNEE1400 pKa = 4.65ADD1402 pKa = 4.18PEE1404 pKa = 4.23IPANRR1409 pKa = 11.84TISTPNPRR1417 pKa = 11.84IGVTVMTGAEE1427 pKa = 4.15TSDD1430 pKa = 3.13QDD1432 pKa = 4.14FGFEE1436 pKa = 3.9ATALQADD1443 pKa = 4.54LSLNKK1448 pKa = 10.14AAFDD1452 pKa = 3.67TSGAALSSAQDD1463 pKa = 3.44GAQIDD1468 pKa = 4.03FVLTVTNDD1476 pKa = 3.44GPATATDD1483 pKa = 3.36VRR1485 pKa = 11.84VRR1487 pKa = 11.84DD1488 pKa = 4.61LIPDD1492 pKa = 4.18GFTYY1496 pKa = 10.7VSNDD1500 pKa = 2.83AAAQAMGYY1508 pKa = 10.06DD1509 pKa = 4.0PSSGIWTLTDD1519 pKa = 3.87LPDD1522 pKa = 3.88GTSATLTLRR1531 pKa = 11.84VVMKK1535 pKa = 10.1ATGTHH1540 pKa = 5.99TNTAEE1545 pKa = 4.16IVASAIPDD1553 pKa = 4.14PDD1555 pKa = 3.69SDD1557 pKa = 4.16PAVGTLRR1564 pKa = 11.84DD1565 pKa = 4.19DD1566 pKa = 4.17LSDD1569 pKa = 4.93GIVDD1573 pKa = 4.55DD1574 pKa = 5.49DD1575 pKa = 4.11EE1576 pKa = 7.04ASATVSFIGMGPTLSGTVFRR1596 pKa = 11.84DD1597 pKa = 3.0NGAAGDD1603 pKa = 3.73TAYY1606 pKa = 10.77DD1607 pKa = 3.95GLQGAQEE1614 pKa = 4.16PGIPDD1619 pKa = 3.35ATVTVADD1626 pKa = 3.78TVGTVLGQPVVAADD1640 pKa = 4.13GTWSLTLPEE1649 pKa = 4.85GFNSAVTVTLAPTSDD1664 pKa = 3.39GQVVSEE1670 pKa = 4.99SPGTLPGLVNTQARR1684 pKa = 11.84DD1685 pKa = 3.29GSFGFTPVSGGTYY1698 pKa = 9.86SNLNFGVIPSARR1710 pKa = 11.84LRR1712 pKa = 11.84NSQQAAMRR1720 pKa = 11.84SGQVITLRR1728 pKa = 11.84HH1729 pKa = 6.5EE1730 pKa = 4.29YY1731 pKa = 8.85TAQAPGNVTFEE1742 pKa = 4.33IDD1744 pKa = 3.28VHH1746 pKa = 5.35QATMPGLFSTGIFLDD1761 pKa = 4.29PACNGTPITAVNGAQPITADD1781 pKa = 3.3TRR1783 pKa = 11.84ICLVIRR1789 pKa = 11.84VTASSAASPGASIAFDD1805 pKa = 4.86LIADD1809 pKa = 4.13TTYY1812 pKa = 11.4GATGLSAQVRR1822 pKa = 11.84NTDD1825 pKa = 3.2VVRR1828 pKa = 11.84VDD1830 pKa = 3.53SSQGQLTLRR1839 pKa = 11.84KK1840 pKa = 7.12TVRR1843 pKa = 11.84NITQGTPEE1851 pKa = 4.27GVSNGAALGDD1861 pKa = 3.56VLEE1864 pKa = 4.03YY1865 pKa = 10.38RR1866 pKa = 11.84IYY1868 pKa = 10.99LDD1870 pKa = 4.36NPGTLPATQIVIHH1883 pKa = 7.11DD1884 pKa = 4.06RR1885 pKa = 11.84TPPYY1889 pKa = 7.57TTLAAPIPSSTAIGTDD1905 pKa = 3.87VICTLAVPSPNNAGYY1920 pKa = 10.53AGPLRR1925 pKa = 11.84WEE1927 pKa = 4.7CAGTHH1932 pKa = 6.22PPGATGSVTFQVQIAPP1948 pKa = 3.74
Molecular weight: 202.22 kDa
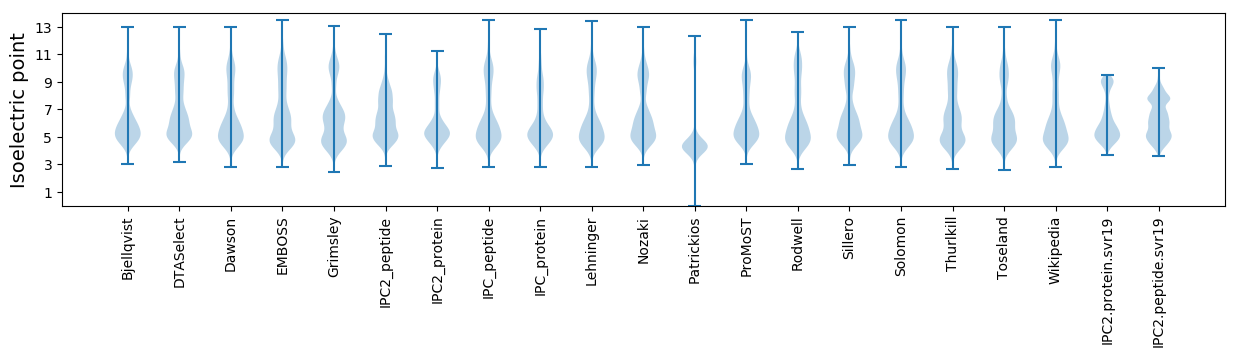
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1C2H9L7|A0A1C2H9L7_9RHOB Uncharacterized protein OS=Thioclava sp. SK-1 OX=1889770 GN=BFP70_17735 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.12GGRR28 pKa = 11.84RR29 pKa = 11.84VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.0GRR39 pKa = 11.84KK40 pKa = 8.91SLSAA44 pKa = 3.86
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.12GGRR28 pKa = 11.84RR29 pKa = 11.84VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.0GRR39 pKa = 11.84KK40 pKa = 8.91SLSAA44 pKa = 3.86
Molecular weight: 5.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1238436 |
32 |
17860 |
324.3 |
35.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.189 ± 0.055 | 0.928 ± 0.017 |
6.188 ± 0.051 | 5.246 ± 0.054 |
3.641 ± 0.037 | 8.483 ± 0.064 |
2.127 ± 0.026 | 5.412 ± 0.027 |
3.149 ± 0.044 | 10.045 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.878 ± 0.03 | 2.668 ± 0.023 |
5.028 ± 0.045 | 3.827 ± 0.031 |
6.399 ± 0.052 | 5.388 ± 0.036 |
5.697 ± 0.065 | 7.125 ± 0.036 |
1.367 ± 0.018 | 2.215 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
